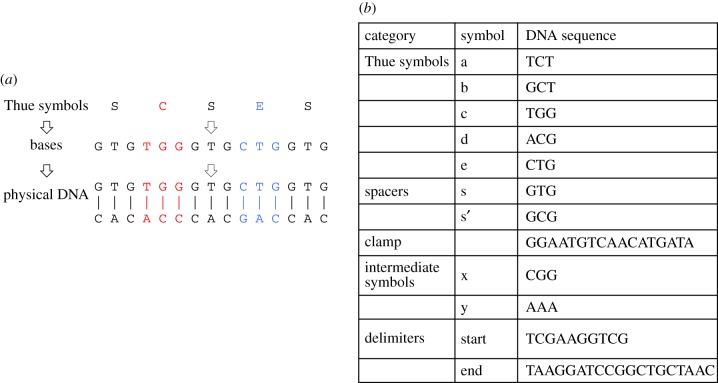
Figure 3.
DNA computing. (a) Three levels of symbol exist in our NUTM: physical DNA, bases, and Thue symbols. Note that molecular structure of single-stranded DNA is asymmetric: one end is termed 3′, the other 5′ (these terms refer to the connectivity of the ribose sugars). When double-stranded DNA is formed the single strands bind together in an anti-parallel manner. DNA polymerases (enzymes for copying DNA) can only copy DNA in the 5′ to 3′ direction. (b) We encode the five Thue system symbols a, b, c, d, e using triplets of DNA. This length was found to provide the best balance between symbol specificity and the ability to mismatch—intriguingly the ‘genetic code’ (cypher) is also based on triplets. The use of triplets helps ensure that when performing PCRs with an annealing temperature in the range of 50–60°C only the desired target sequences are amplified. A spacer symbol s (or s') occurs between each Thue symbols to help enforce the specificity of desired mismatches. We require the marker symbol clamp for rule recognition. We use the intermediate symbols x and y to help ensure that unwanted cross-hybridization of symbols does not occur. Finally, the symbols start and end delimit the program. Physically, these delimiters are used as recognition site primers for PCRs.