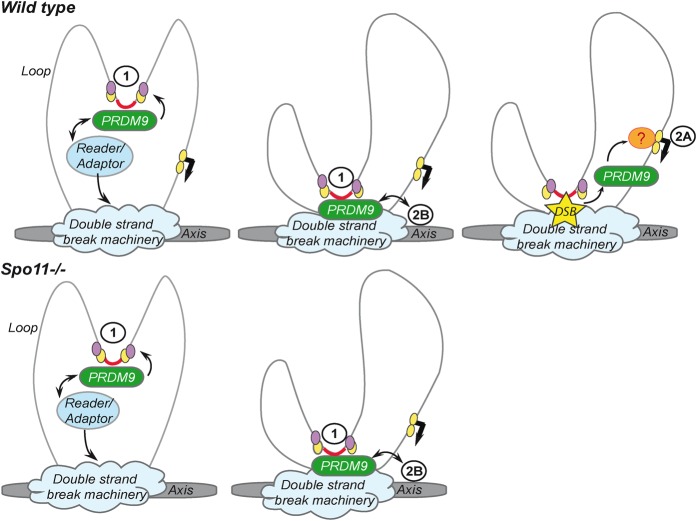
Figure 6.
Model of PRDM9 binding dynamics in mouse testes. In early meiotic prophase, when PRDM9 is expressed, chromosomes are predicted to be organized in a characteristic loop-axis structure, shaped by cohesins and other proteins. Some essential components of meiotic DSB formation, such as MEI4 and IHO1, are located as discrete foci on the axis. In wild-type animals, PRDM9, through its zinc finger domain, binds to class 1 DNA motifs (red) that are meiotic recombination hotspots and located in chromatin loops. PRDM9 modifies the surrounding nucleosomes by promoting H3K4me3 (yellow) and H3K36me3 (purple) deposition. Then, a reader or adaptor protein promotes PRDM9 interaction with the DNA double-strand break formation machinery located on the axis. Therefore, PRDM9 can also indirectly interact with DNA sequences near or on the axis. We call these sites PRDM9 class 2B binding sites. SPO11 may be recruited at PRDM9 binding sites before or after the loop-axis interaction. Upon DSB formation, PRDM9 could be displaced and interact with other sites (class 2A that are mainly transcription start sites), possibly through interaction with other unknown factors (question mark). Class 2 sites differ in mouse strains with different PRDM9 alleles. The selection of class 2 sites may depend on their spatial proximity to class 1 sites. In mice deficient for SPO11 (Spo11−/− or Spo11KO), PRDM9 binds to class 1 sites and is brought close to the axis, leading to interactions detected as class 2B sites. However, in the absence of SPO11, PRDM9 is not competent or not released in a timely manner to bind to class 2A sites.