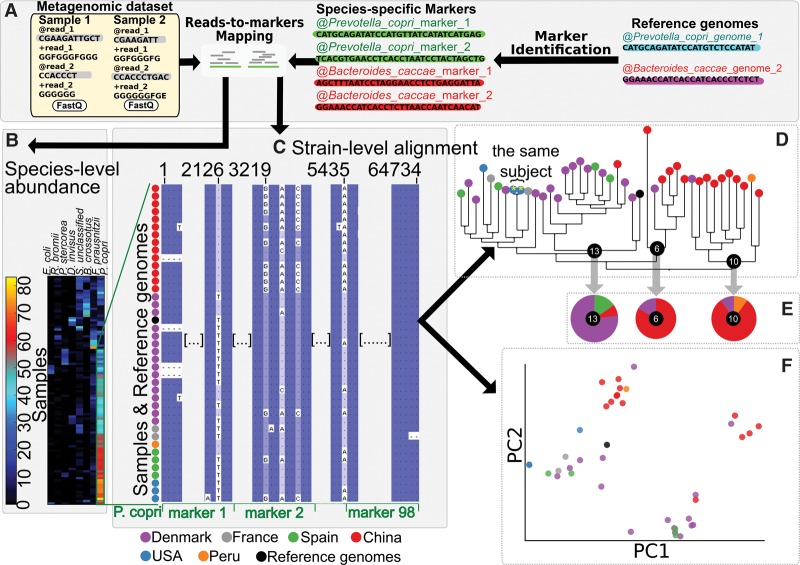
Figure 1.
StrainPhlAn for strain identification and tracking in shotgun metagenomes and its application to Prevotella copri in the human gut. StrainPhlAn provides a method to identify strains from shotgun metagenomes and provides tracking, comparative, and phylogenetic analyses across samples. Here, we illustrate results using Prevotella copri as an example species in a demonstration subset of this study's human gut metagenomes. (A) In this overview of the method, for each species for which strains are to be analyzed across a metagenome collection, sample-specific and strain-specific markers are constructed by mapping reads against the MetaPhlAn2 (Truong et al. 2015) database of species-specific reference sequences. (B) In each sample, species are identified and quantified if sufficient coverage for the species markers is detected. Here, 100 samples with sufficiently abundant P. copri are shown (seven other abundant species are also displayed). (C) The preselected species-specific markers are concatenated, aligned, and variants identified using the consensus sequence of mapped metagenomic reads. (D) From the resulting set of the most abundant strains per sample, a phylogenetic tree can be built. This allows, for example, retained or minimally divergent strains within a particular environment (e.g., human host) to be easily identified when they appear within the same subtrees. (E) Strains or subtrees can also be statistically associated with sample metadata (e.g., human or environmental phenotypes). (F) Each species’ genetic diversity and divergence can be easily visualized as an ordination comparable to those used for isolate or human population genetics.