Figure 1.
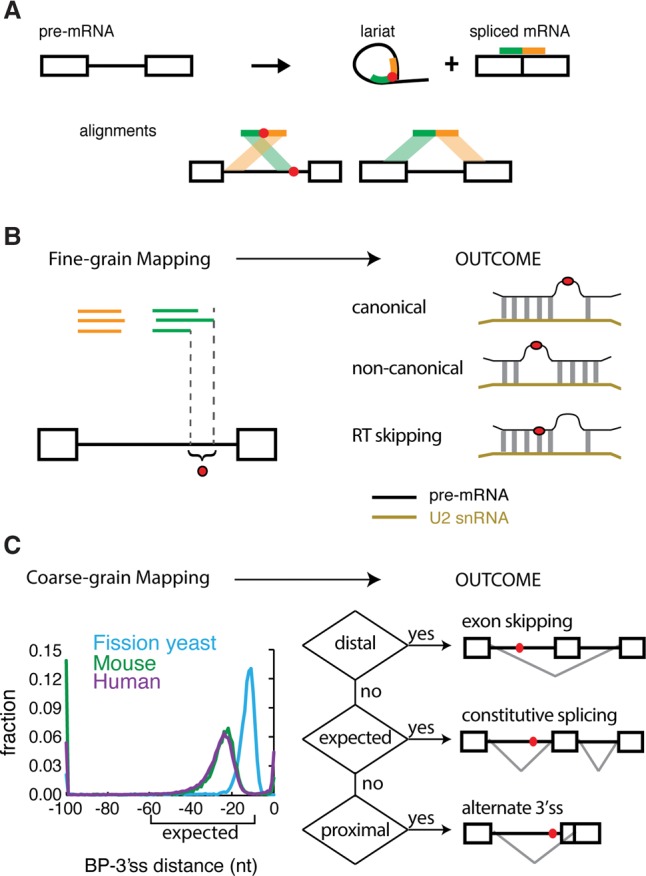
The technique and purpose of mapping branchpoints. (A) Overview of branchpoint mapping (adapted by permission from Macmillan Publishers Ltd: [Nature Structural and Molecular Biology] (Taggart et al. 2012), © 2012). Inverted fragmental cDNA reads across the 2′-5′ linkage of the lariat RNAs were collected to map the branchpoint. (B) Fine-grain mapping. Discovering the precise locations of branch sites enables the discovery of canonical and noncanonical modes of U2snRNA pairing and the characterization of RT ‘skipping’ behavior around 2′-5′ linkages. (C) Coarse-grain mapping. General branchpoint location was analyzed in three species: fission yeast, mouse, and human. The human and mouse branchpoints were largely located within the expected region (between 10 and 60 nt upstream of the 3′ss AG), but the fission yeast branchpoints were more proximal. The ENCODE RNA-seq data sets were used to test the relationship between branchpoint location and alternative splicing outcomes.
