Figure 2.
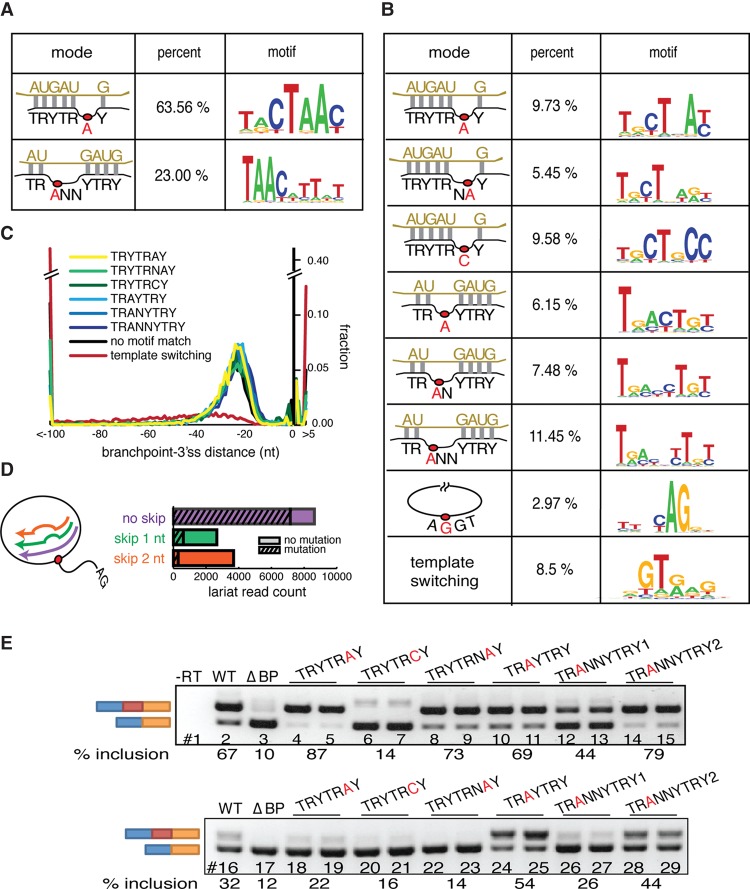
Alternate modes of U2snRNA:pre-mRNA base-pairing emerged from lariat mapping. (A) U2snRNA:pre-mRNA base-pairing models found in Schizosaccharomyces pombe. Twenty-nucleotide windows around unprocessed branchpoint reads were iteratively aligned to U2snRNA in a manner that allowed branchpoint location, loop location, and the loop length to vary. Significantly enriched modes of base-pairing were identified and recorded. Left column: schematics of significantly enriched modes of base-pairing, middle column: percent of lariat reads that fit model, right column: motif of lariat reads that fit model. (B) U2snRNA:pre-mRNA base-pairing models found in human. Left column: schematics of significantly enriched modes of base-pairing, middle column: percent of lariat reads that fit model, right column: motif of lariat reads that fit model. (C) Branch site motif distribution relative to the 3′ss. (D) The skipping behavior and mutational profile of reverse transcriptase (RT) was inferred from the human canonical branchpoint (TRYTRAY) mapping. The histogram describes the frequency of each size of RT-induced skip (x-axis). The striped portion of bars represents the fraction of misincorporation events at the apparent branchpoint. (E) Exemplars of branch site motifs were validated in a strong (APRT, upper) and weak (SHMT2, lower) branch site reporter minigene. Branch sites of the first intron (between the blue and red exons) were first deleted (ΔBP, lanes 3,17) or replaced by branch site motifs identified in B to test for their ability to restore splicing. Inclusion level of the middle exon is indicated under the gel image.