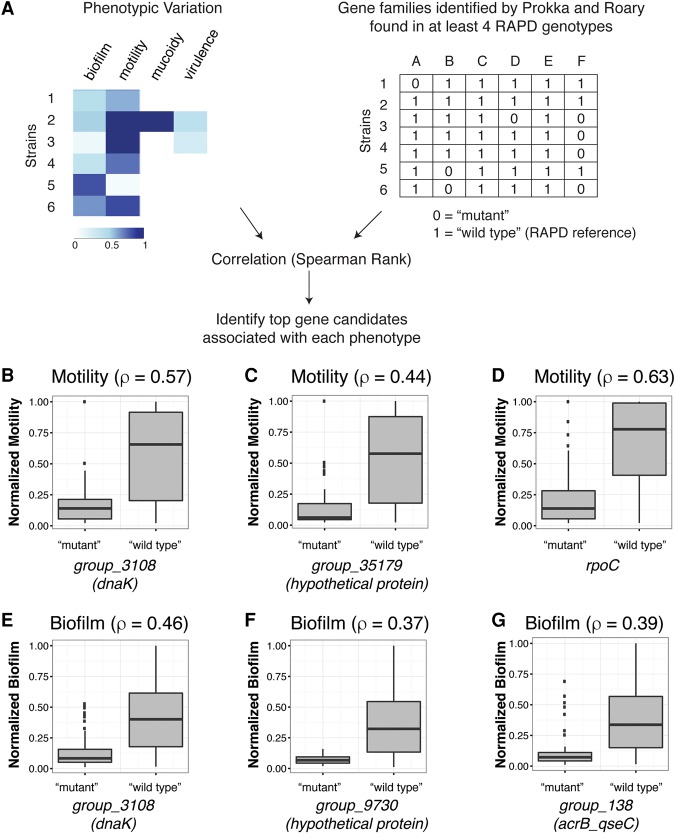
Figure 7.
Genetic association testing identifies candidate gene families for the observed phenotypic variation. (A) We generated two matrices, one representing the phenotypic variation and the other the genotypic variation where we defined the variation for each isolate with respect to its RAPD reference, with 1 = “wild type” and 0 = “mutant,” i.e., any allele affected by deletion, nonsense, or missense mutations compared with the appropriate RAPD PacBio reference sequence from the earliest time point. Spearman rank correlation found genes that were highly correlated with either motility (examples in B–D) or biofilm (examples in E–G).