FIGURE 4.
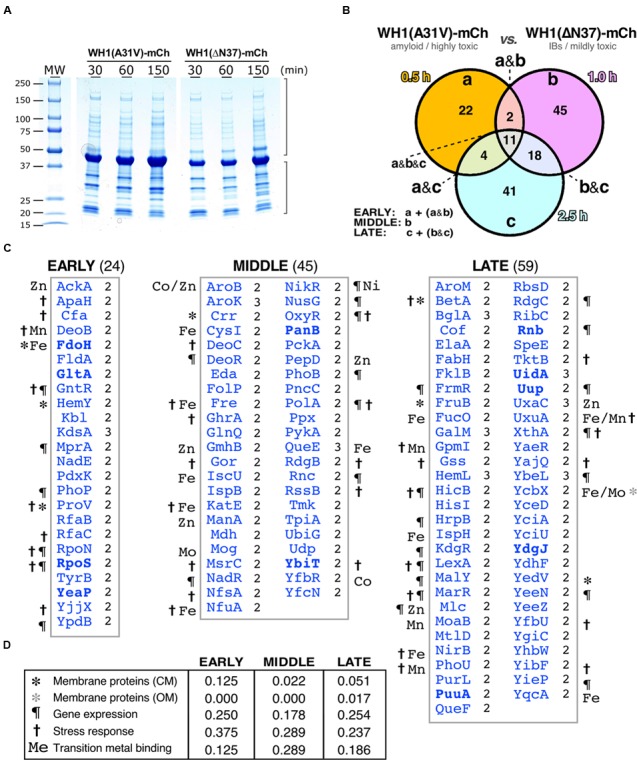
Differential interactomics in E. coli cells expressing the A31V or the ΔN37 mutants of RepA-WH1. (A) SDS-PAGE showing the aggregated protein fraction from bacteria expressing either WH1(A31V)-mCh or the WH1(ΔN37)-mCh mutant. (B) Venn diagram displaying the temporal distribution of the proteins found exclusively co-aggregated with the A31V variant of the prionoid. (C) Lists of the proteins found co-aggregated with WH1(A31V)-mCh, but not with ΔN37, in at least two of the three biological replicas (right-hand notation: 2/3; Supplementary Dataset S2) at the indicated time slots (B). Symbols correspond to the GO terms, as described in panel D (to compare with transcriptomics, see Figure 3C). The eight proteins in common with the transcriptomic dataset (Figure 3) are in boldface. (D) Temporal distribution across five main functional gene ontology (GO) terms of the proteins preferentially co-aggregated with WH1(A31V)-mCh.
