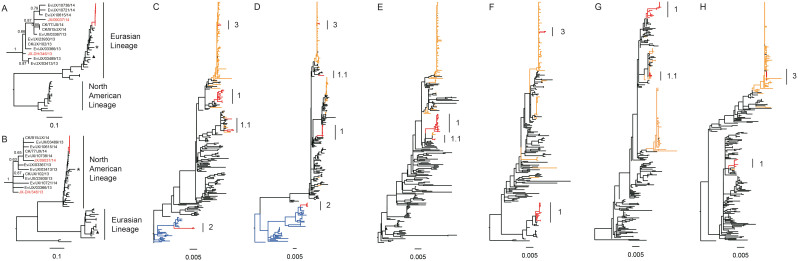
Figure 3. Phylogenetic analyses for eight gene segments of H10N8 viruses.
The maximum likelihood tree for each segment, including HA (A), NA (B), PB2(C), PB1 (D), PA (E), NP (F), MP (G) and NS(H) of the novel H10N8 influenza viruses is shown. The sequences for novel H10N8 viruses and H9N2 viruses in the study, novel H7N9 viruses, other H9N2 viruses and wild birds gene pool are highlighted in red, orange, black and blue, respectively. The major group of H9N2 is labeled as Clade 1, and the minor group is denoted as 1.1 (labeled with a black line). The groups within the duck H7Nx clade are shown as a light blue line and labeled as Clade 2, and the groups within the major H7N9 clade are shown as a yellow line and labeled as Clade 3. Two previous H10N8 viruses, A/environment/DongtingLake/Hunan/3–9/2007 and A/duck/Guangdong/E1/2012, are denoted as a triangle and star, respectively. The phylogenic tree of HA and NA of H10N8 viruses in the study was zoomed out and the name of H10N8 viruses from human or non-human was denoted in red and black, respectively. The sequences of other viruses used for the analysis were downloaded from the Global Initiative on Sharing All Influenza Data (GISAID) database and are shown in black. The regions of the nucleotide sequences used for the phylogenetic analysis were as follows: H10, 1,686 bp; N8, 1,413 bp; PB2, 2,280 bp; PB1, 2,277 bp; PA, 2,151 bp; NP, 1,497 bp; MP, 982 bp and NS, 838 bp.