Figure 5.
DCs from Infarcted Heart Have an Activated Phenotype
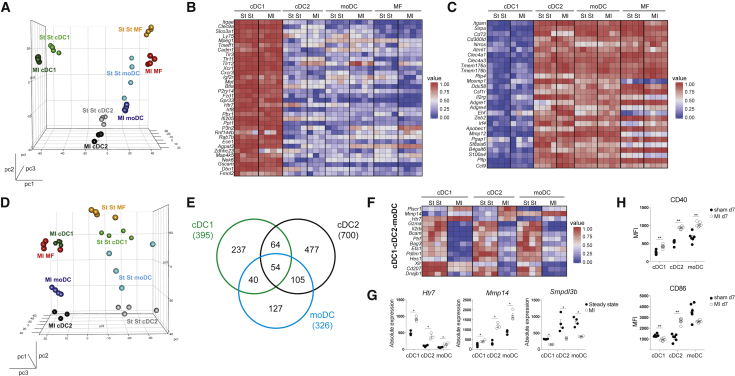
(A) Front view of PCA of RNA-seq data from cDC1s, cDC2s, moDCs, and MFs in the steady-state (St St) and the infarcted (MI d7) heart. PCA was calculated using the top 15% most varying genes between cell subsets. Each dot symbolizes one independently sorted replicate of the indicated cell population, and four independent sorts were performed per subset.
(B and C) Heat map of relative expression of hallmark cDC1 genes (B) and hallmark cDC2 genes (C) in cDC1s, cDC2s, moDCs, and MFs sorted from steady-state and infarcted hearts.
(D) Side view of PCA of RNA-seq data from cDC1s, cDC2s, moDCs, and MFs in the steady-state and infarcted heart.
(E) Venn diagram showing numbers and overlap of DEGs in cDC1s, cDC2s, and moDCs sorted from MI d7 compared to steady-state hearts.
(F) Heat map of relative expression of top-shared DEGs between DC subsets from MI d7 hearts compared to corresponding subsets isolated from the steady-state heart. To calculate top DEGs, a threshold of a minimum 1.5 Log2 fold change was used. As an exception, no threshold was set on shared upregulated genes of cDC1s, cDC2s, and moDCs.
(G) Bar graphs representing absolute expression of DEGs among cDC1s, cDC2s, and moDCs sorted from steady-state and infarcted hearts.
(H) MFI of CD40 and CD86 expression (day 7 post-surgery) on heart DC subsets in sham-operated compared to the infarcted heart (n = 6). All bar graphs in Figure 5 show data as mean ± SEM (∗p ≤ 0.05; ∗∗p ≤ 0.01), and all data are representative of four independent experiments.
See also Figure S1 and Tables S1–S7.