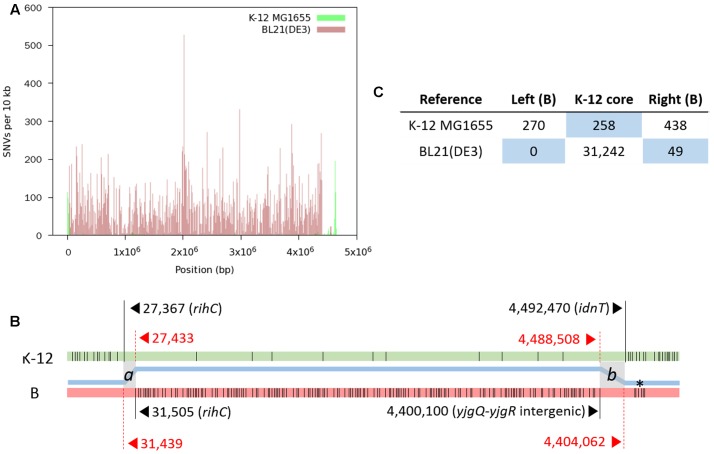
FIGURE 2.
Distribution of SNVs identified by mapping RR1 reads to reference genomes. (A) SNV density plot across each reference genome. (B) A schematic representation of SNVs (thin vertical lines) on each reference genome (not drawn to scale). For each block of SNVs representing the heterologous genomic segments, the first and last SNVs are denoted by long vertical lines with coordinates and gene symbols. The thick blue line between the two genomes depicts the hybrid genome structure resulting from recombination between the K-12 (green) and B (pink) genomes. Two crossover events took place at the gray zones (a and b), where the two parental genomes are identical to each other. Locus tags of RR1 for rihC, yjgQ, yjgR, and idnT are SR35_00165, SR35_21895, SR35_21900, and SR35_21910, respectively. An asterisk denotes the K-12 island in the B region. (C) Total numbers of SNVs in each block on the reference genomes. The number of SNVs in the right B-block includes one SNV within the b crossover region. The blue background emphasizes recombinational blocks within the parental genomes.