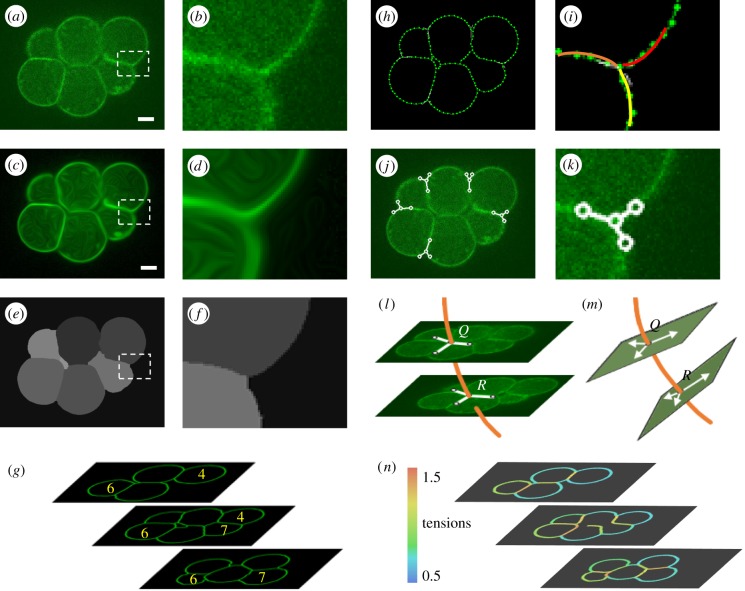
Figure 1.
An overview of CellFIT-3D. A raw confocal image of an eight-cell stage murine embryo is shown in (a) (scale bar, 10 µm), while (b) shows an enlargement of the boxed area. An edge enhancing coherence filter [39] was used to smooth the boundaries (c) and close their gaps (d). Parameters used: Scheme, implicit discretization; total diffusion time = 25; Gaussian sigma = 3. SeedWater [40]-segmented cells are visible in (e) and (f), each denoted by in a different intensity of grey. Corresponding cells in successive images were grouped together (g) and TEs that appeared in multiple images identified. A fine mesh (h) was constructed along each cell boundary and its approach angle to any given triplet determined by circular arc fitting (i). Tangent vectors (j) were calculated automatically and adjusted manually as needed (k). Splines were fit through the triplets that appeared in successive images (l), and dihedral planes and angles calculated (m). Least-squares equations were constructed and solved (see §2i,j) and calculated tensions displayed (n).