Fig. 2.
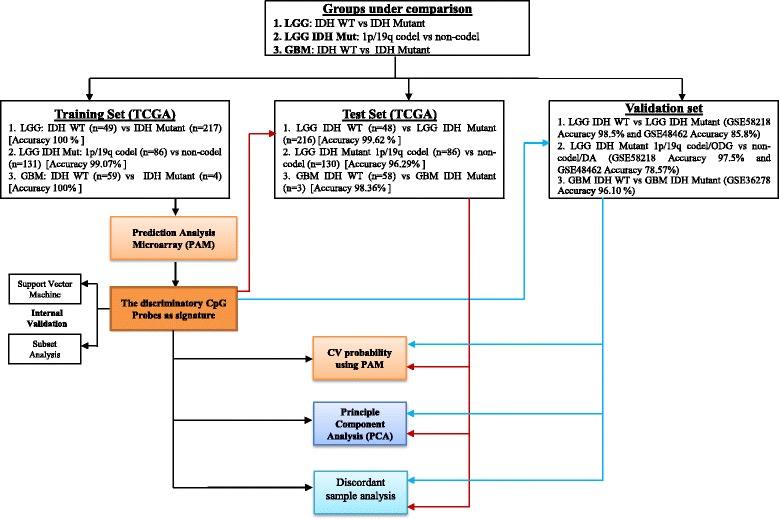
The schematic representation of the work flow of statistical analysis. PAM identified 14 discriminatory CpG probes of DNA methylation between (1) IDH Mut (LGG IDH Mut) and WT (LGG IDH WT) which was further validated by principal component analysis (PCA). Fourteen CpG probe methylation signatures were then validated in test set. Here, TCGA dataset (450K methylation) was randomly divided into equal halves to form the training and test set. Similar protocol was performed for (2) LGG IDH Mut 1p/19q intact (diffuse astrocytoma/DA) versus LGG IDH Mut 1p/19q codel (oligodendroglioma/ODG) and (3) GBM IDH Mut versus WT. All the derived methylation signatures are validated in independent validation datasets with high accuracy
