Fig. 7.
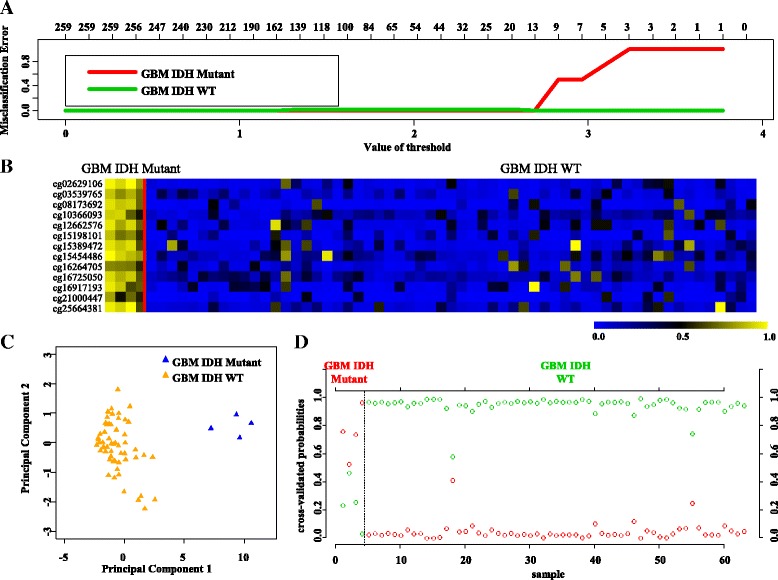
Identification of 13 CpG probe methylation signatures in training set (TCGA) for IDH Mut and WT in GBM. a Plot demonstrating classification error for 259 CpG probes from PAM analysis in training set. The threshold value of 2.694 corresponded to 13 discriminatory CpG probes which classified IDH Mut (n = 4) and WT (n = 59) GBM samples with classification error of 0%. b Heat map of the 13 CpG discriminatory probes identified from the PAM analysis between IDH Mut and WT GBM patient samples in the training set (TCGA). A dual color code was used where yellow indicates more methylation (hypermethylation) and blue indicates less methylation (hypomethylation). c PCA was performed using beta (methylation) values of 13 PAM-identified CpG probes between IDH Mut (n = 4) and WT (n = 59) GBM samples in training set. A scatter plot is generated using the first two principal components for each sample. The color code of the samples is as indicated. d The detailed cross-validation probabilities of 10-fold cross-validation for the samples of training set based on the beta values of 14 CpG probes are shown. For each sample, its probability as IDH Mut (red color) and WT (green color) GBM samples is shown and it was predicted by the PAM program as either IDH Mut or WT in GBM samples based on which grade’s probability is higher. The original histological grade of the samples is shown on the top
