Fig. 1.
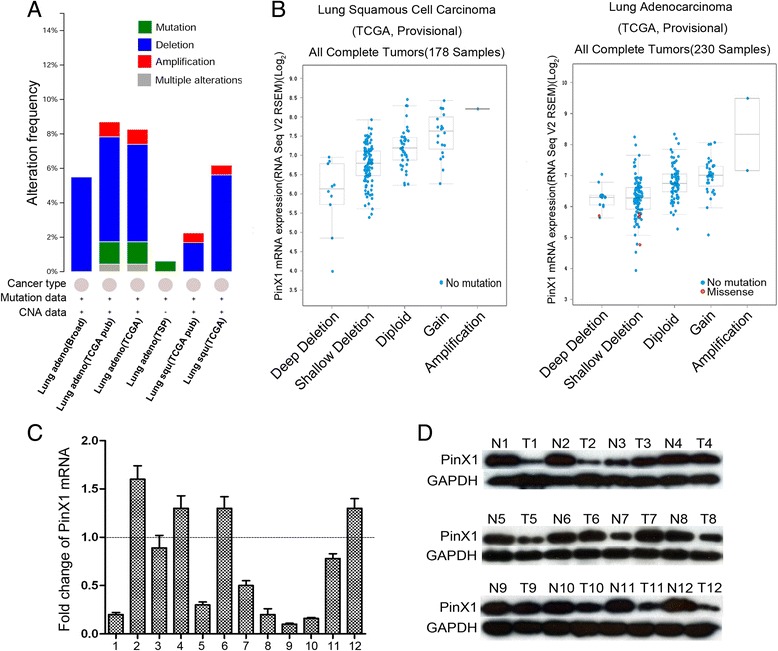
The alterations of PinX1 gene in cBioportal Web resource online and PinX1 expression in 12 primary NSCLC tissues. a The alterations of PinX1 gene were explored in six NSCLC database (Lung Adenocarcinoma – Broad, Cell 2012; TCGA, Provisional; TCGA, Nature, 2014; TSP, Nature 2008; Lung Squamous Carcinoma – TCGA, Provisional; TCGA, Nature 2012). b Relative expression level as a function of relative copy number of PinX1 genes were plotted in two NSCLC database respectively (Lung Squamous Carcinoma – TCGA, Provisional; Lung Adenocarcinoma –TCGA, Provisional). Deep Deletion — homozygously deleted; Shallow Deletion — heterozygously deleted; Diploid — two alleles present; Gain — low-level gene amplification event; Amplification — high-level gene amplification event. c Decreased expression of PinX1 mRNA was examined by qRT-PCR in 8/12 NSCLC cases, when compared with adjacent normal lung tissues. Expression levels were normalized for GAPDH. These samples (12 NSCLC samples) were randomly picked from the learning cohort. Error bars, SD, calculated from three parallel experiments. d Decreased expression of PinX1 protein was detected by western blotting in 7/12 NSCLC cases, when compared with adjacent normal lung tissues. These samples (12 NSCLC samples) were randomly picked from the learning cohort. N, adjacent normal lung tissues; T, primary NSCLC samples
