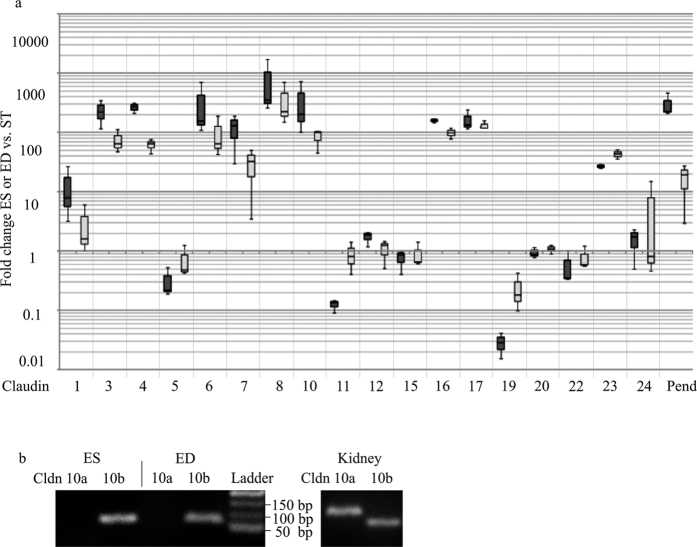
Figure 3. Claudin mRNA expression in rat p4 ED and ES epithelium.
(a) High throughput qPCR analysis of ED and ES mRNA. TaqMan probes against exon-exon boundaries of all mRNA subtypes were used (Table 1). Box-whisker-blot shows 25–75 percentile of the normalized relative quantities (NRQs) of rat p4 ED (n = 3; light gray bars) or ES (n = 3; black bars) claudin mRNA expression versus claudin mRNA expression of surrounding tissue (ST). Claudin 3, 4, 6, 7, 8, 10, 16, 17 and 23 mRNA were found to be expressed considerable stronger in the ED and ES with a NRQ of 10 or more if compared with surrounding tissue (ST). Claudins not shown were not expressed in the ES or ED (n = 3). Positive control target: Pendrin (Pen). (b) Cropped gel images of RT-PCR analysis of the claudin 10a and 10b mRNA isoforms for rat p4 ED and ES epithelium. Only Claudin 10a transcripts were obtained, whereas the Claudin 10b primers yielded no transcript in ED and ES. Both transcripts, Claudin 10a and 10b, were obtained in the positive control tissue of rat p4 kidney. DNA Ladder: 50 bp.