Fig. 2.
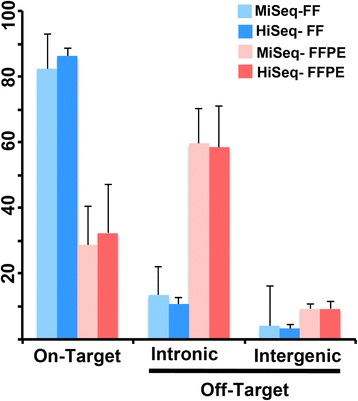
MiSeq and HiSeq platform mapped read comparison from FF- and FFPE-derived RNA sequences. a Barplot depicts the percentage of mapped reads that are on-target, or off-target (intronic and intergenic) for FF and FFPE samples processed on MiSeq and HiSeq platforms. b Beeswarm box plot shows mapped reads (%) form individual FF (blue) and FFPE (red) samples processed on the HiSeq
