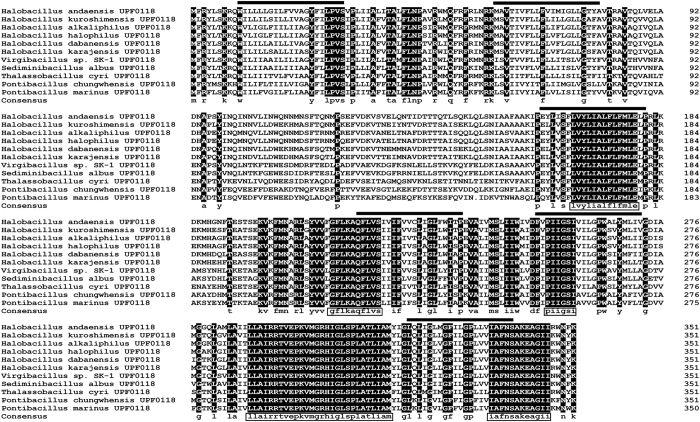
Figure 2. Alignment of UPF0118 with its most closely related holomogs clustered within the neighbour-joining phylogenetic tree.
The ten closest homologs with 58–82% identities clustered within the neighbour-joining phylogenetic tree with the bootstrap value of 100% (Fig. 3) were selected to show the conserved motifs and amino acid residues. Acession.version numbers are shown in the neighbour-joining phylogenetic tree of Fig. 3. Shading homology corresponds to 100% (black), >75% (grey), ≥50% (lightgrey) and <50% (white) amino acid identity, respectively. The six putative transmembrane segments are marked with bold lines above the alignment. The five highly conserved motifs are highlighted with the open rectangles in the consensus sequence.