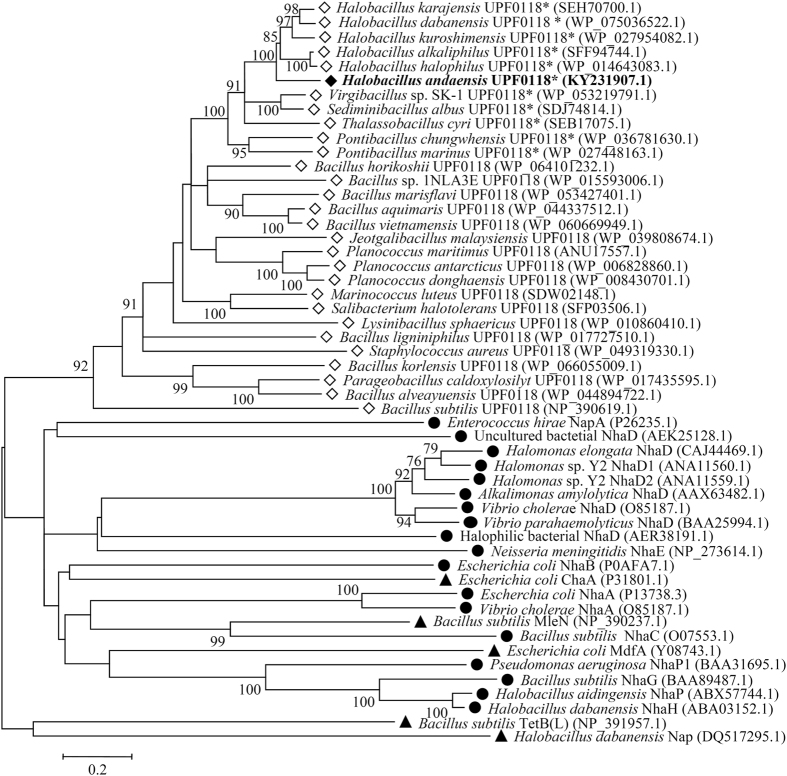
Figure 3. Neighbour-joining phylogenetic tree of selected homologs of Halobacillus andaensis UPF0118, together with known Na+/H+ antiporters.
For the construction of phylogenetic tree, nine closest homologs with 60–82% identities, nine closer homologs with 49–59% identities, nine distant homologs with 22–42% identities, together with all representatives of known specific single-gene Na+/H+ antiporters and single-gene proteins with the Na+/H+ antiport activity were selected. Accession.version numbers of selected homologs were shown in the parenthesis. Open diamond stands for putative UPF0118 family proteins; filled diamond stands for UPF0118 identified in this study; filled circle stands for known specific single-gene Na+/H+ antiporters; filled triangle stands for other single-gene proteins with the Na+/H+ antiport activity. Bootstrap values ≥70% (based on 1000 replications) are shown at branch points. Bar, 0.2 substitutions per amino acid residue position. UPF0118 and its most closely related holomogs clustered with the bootstrap value of 99% marked with the asterisk were used for protein alignment in Fig. 2.