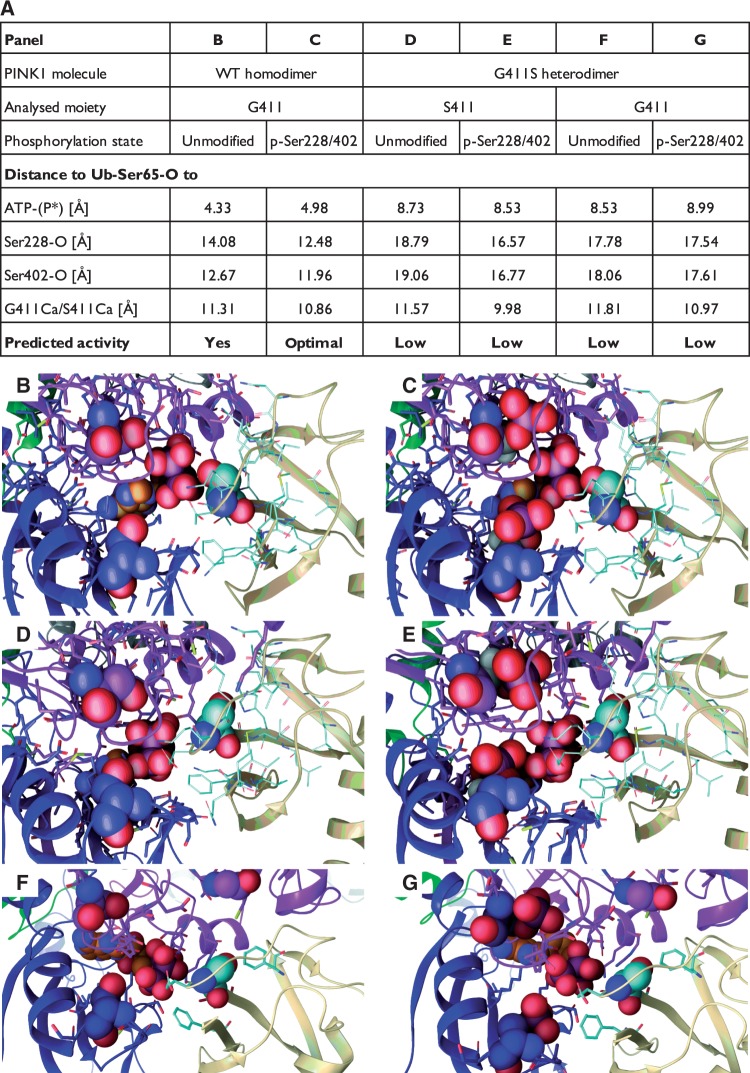
Figure 6.
Ubiquitin binding to PINK1 and prediction of phosphorylation efficiencies. Ubiquitin was docked as a substrate to each half of the PINK1 kinase wild-type homo- (G411/G411) and mutant (G411/S411) heterodimers. Given is a tabular summary of distances [Å] between the Ser65 oxygen of ubiquitin (Ub-Ser65-O) and the terminal phosphate of the bound ATP molecule as well as key PINK1 atoms (A). Relevant PINK1 atoms include the oxygens of Ser228 and Ser402 (Ser228/402-O) as well as the alpha carbons of either G411 or S411 (G411/S411-Cα). Both unmodified and auto-phosphorylated (p-Ser228/402) forms of PINK1 were analysed and compared for each subunit from the respective dimers. Greater distances between Ub-Ser65 and ATP as well as Ser228 and Ser402 of PINK1 in both wild-type and mutant subunits of the heterodimer, compared to wild-type homodimer, likely result in less efficient phosphorylation of the substrate. Interestingly, phosphorylation of PINK1 Ser228 and Ser402 in the wild-type homodimer facilitates an optimal alignment of ATP and Ub-Ser65 in the active site of PINK1. (B–G) The corresponding magnifications of the kinase domain (between the N- and C-lobes) of the PINK1 molecules with ubiquitin docked near the active site. Ser228 and Ser402 (or p-Ser228 and p-Ser402) are shown in Van der Waals spheres with the carbons coloured to match the domain colours. ATP is shown with orange carbons and structure in Van der Waals. The ubiquitin molecule is shown in beige ribbons with cyan carbon and the Ser65 residue in Van der Waals. G411 or S411 of PINK1 is shown above the site. Corresponding full-length PINK1 dimer structures in complex with ubiquitin can be found in Supplementary Fig. 6. A schematic representation of the analysed distances between the respective atoms is depicted in Supplementary Fig. 7.