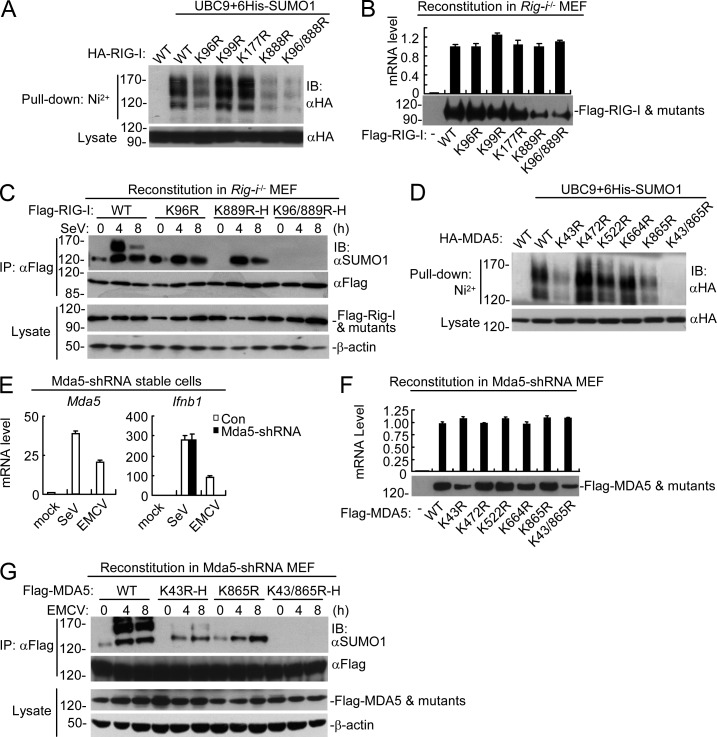
Figure 4.
Identification of sumoylation sites of RIG-I and MDA5. (A) Identification of sumoylation residues of RIG-I. HEK293 cells were transfected with the indicated plasmids for 24 h, followed by Ni2+ pull-down assays and immunoblotting analysis. (B) Expression of RIG-I and its mutants in reconstituted Rig-i−/− MEFs. Reconstitution of RIG-I or its mutants into Rig-i−/− MEFs, followed by analysis of their mRNA and protein levels. (C) Sumoylation of RIG-I and its mutants in reconstituted cells. Rig-i−/− MEFs reconstituted with RIG-I or its mutants were left uninfected or infected with SeV for the indicated times before immunoprecipitation and immunoblotting analysis. To ensure comparable protein levels of RIG-I and its mutants in all reconstituted MEFs, the titers of retroviruses were adjusted for reconstitution. More retroviruses containing RIG-I K889R or K96/889R mutants were used to superinfect the cells, which were designated as K889R-H and K96/889R-H, respectively. (D) Identification of sumoylation residues of MDA5. HEK293 cells were transfected with the indicated plasmids for 24 h, followed by Ni2+ pull-down assays and immunoblotting analysis. (E) Knockdown efficiency of MDA5-shRNA in stably transduced MEFs. The cells were left uninfected or infected with EMCV or SeV for 6 h before qPCR analysis. (F) Expression of MDA5 and its mutants in reconstituted MDA5-shRNA MEFs. Reconstitution of MDA5 and its mutants into MDA5-shRNA MEFs, followed by analysis of their mRNA and protein levels. (G) Sumoylation of MDA5 and its mutants in reconstituted cells. MDA5-shRNA MEFs reconstituted with MDA5 or its mutants were left uninfected or infected with EMCV for the indicated times before immunoprecipitation and immunoblotting analysis. To ensure comparable protein levels of MDA5 and its mutants in all reconstituted MEFs, the titers of retroviruses were adjusted for reconstitution. More retroviruses containing the K43R or K43/865R mutants were used to superinfect the cells, which were designated as K43R-H and K43/865R-H, respectively. All the experiments were repeated for three times.