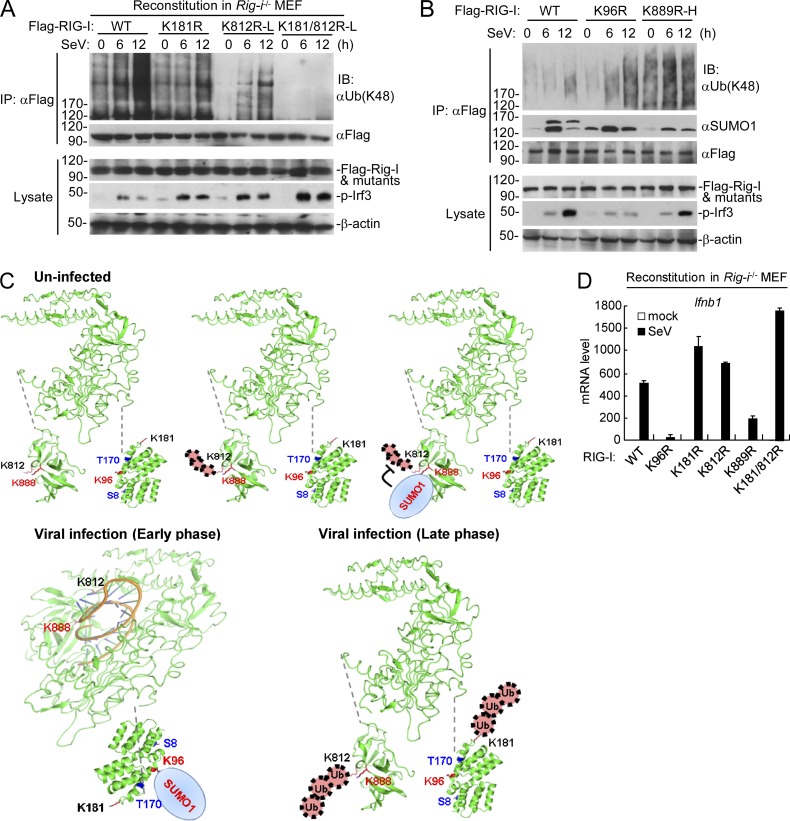
Figure 6.
Physiological relationship of sumoylation and K48-linked polyubiquitination of RIG-I. (A) The K48-linked polyubiquitination of RIG-I and its mutants after SeV stimulation in reconstituted cells. Rig-i−/− MEFs reconstituted with RIG-I, or its mutants were infected with SeV for the indicated times, followed by immunoprecipitation and immunoblotting analysis. As described in Fig. 4 C, subinfection (K812R-L and K812/181R-L) with the RIG-I mutant-containing retroviruses was used to make the reconstituted RIG-I and mutants express at similar levels. (B) Dynamic sumoylation and K48-linked polyubiquitination of RIG-I and its mutants after SeV stimulation in reconstituted cells. Rig-i−/− MEFs reconstituted with RIG-I or its mutants were infected with SeV for the indicated times, followed by immunoprecipitation and immunoblotting analysis (top). As described in C, superinfection the RIG-I K889R mutant (K889R-H) containing retroviruses was used to make the reconstituted RIG-I and its mutants express at similar levels. (C) The schematic diagram for dynamic K48-linked polyubiquitination and sumoylation of RIG-I before and after viral infection. The crystal structures of RIG-I CARD (PDB:4P4H) and Helicase domains (PDB:5E3H) were obtained from the PDB database. Black, ubiquitination sites; red, sumoylation sites; blue, phosphorylation sites. (D) SeV-induced transcription of downstream antiviral genes in Rig-i−/− MEFs reconstituted with wild-type RIG-I or its mutants. The reconstituted cells were left uninfected or infected with SeV for 6 h before qPCR analysis. Data in D are from one representative experiment with three technical replicates (mean ± SD). All the experiments were repeated for three times.