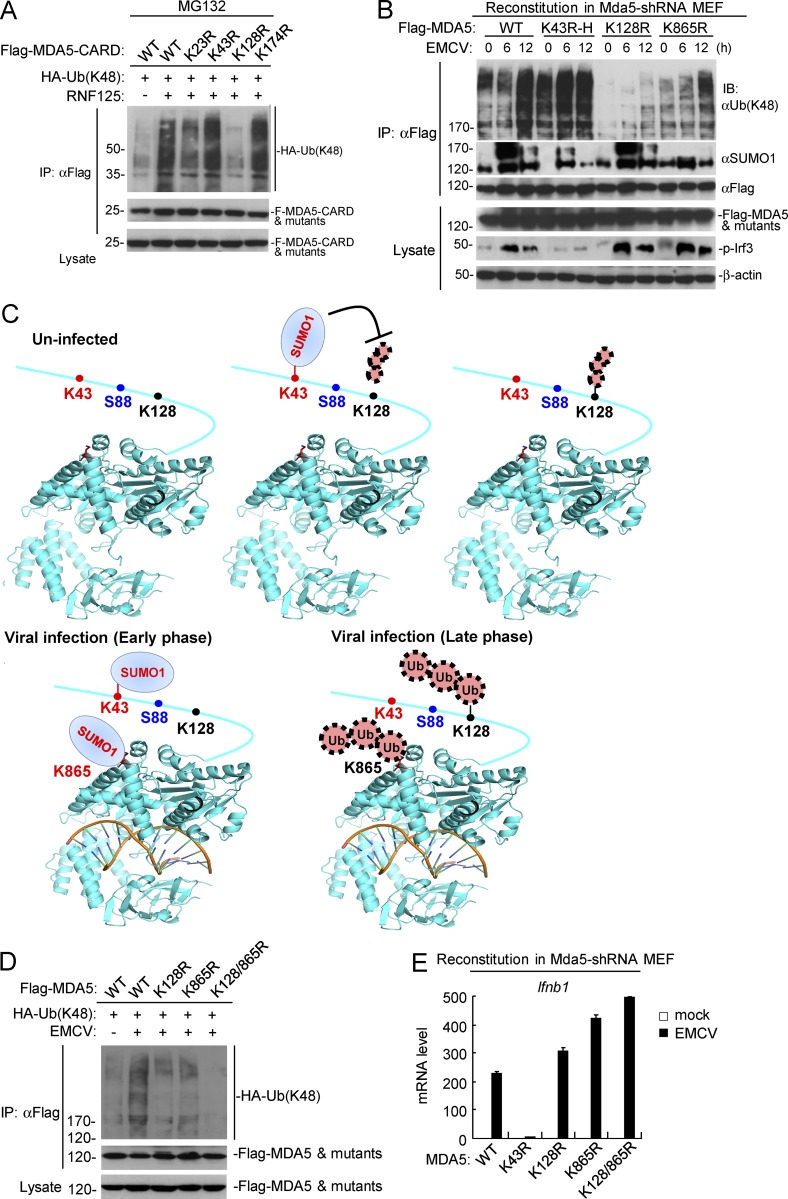
Figure 7.
Physiological relationship of sumoylation and K48-linked polyubiquitination of MDA5. (A) Identification of the residue in MDA5-CARD targeted by RNF125 for K48-linked polyubiquitination. HEK293 cells were transfected with the indicated plasmids for 18 h, and then treated with MG132 for 6 h before immunoprecipitation and immunoblotting analysis. (B) Dynamic sumoylation and K48-linked polyubiquitination of MDA5 and its mutants after EMCV stimulation in reconstituted cells. MDA5-shRNA MEFs reconstituted with MDA5 or its mutants were infected with EMCV for the indicated times, followed by immunoprecipitation and immunoblotting analysis (top histogram). As described in Fig. 4 G, superinfection (K43R-H) or subinfection (K128R-L) of the MDA5 mutant-containing retroviruses was used to make the reconstituted MDA5 and its mutants express at similar levels. (C) The schematic diagram for dynamic K48-linked polyubiquitination and sumoylation of MDA5 before and after viral infection. The crystal structure of MDA5 Helicase domain (PDB: 4GL2) was obtained from the PDB database. Black, ubiquitination sites; red, sumoylation sites; blue, phosphorylation site. (D) Effects of simultaneous mutation of K128 and K865 on EMCV-induced K48-linked polyubiquitination of MDA5. HEK293 cells were transfected with the indicated plasmids for 24 h, and then cells were left uninfected or infected with EMCV for 12 h, followed by immunoprecipitation and immunoblotting analysis. (E) EMCV-induced transcription of downstream antiviral genes in MDA5-shRNA MEFs reconstituted with wild-type MDA5 or its mutants. The reconstituted cells were left uninfected or infected with EMCV for 6 h before qPCR analysis. Data in E are from one representative experiment with three technical replicates (mean ± SD). All the experiments were repeated three times.