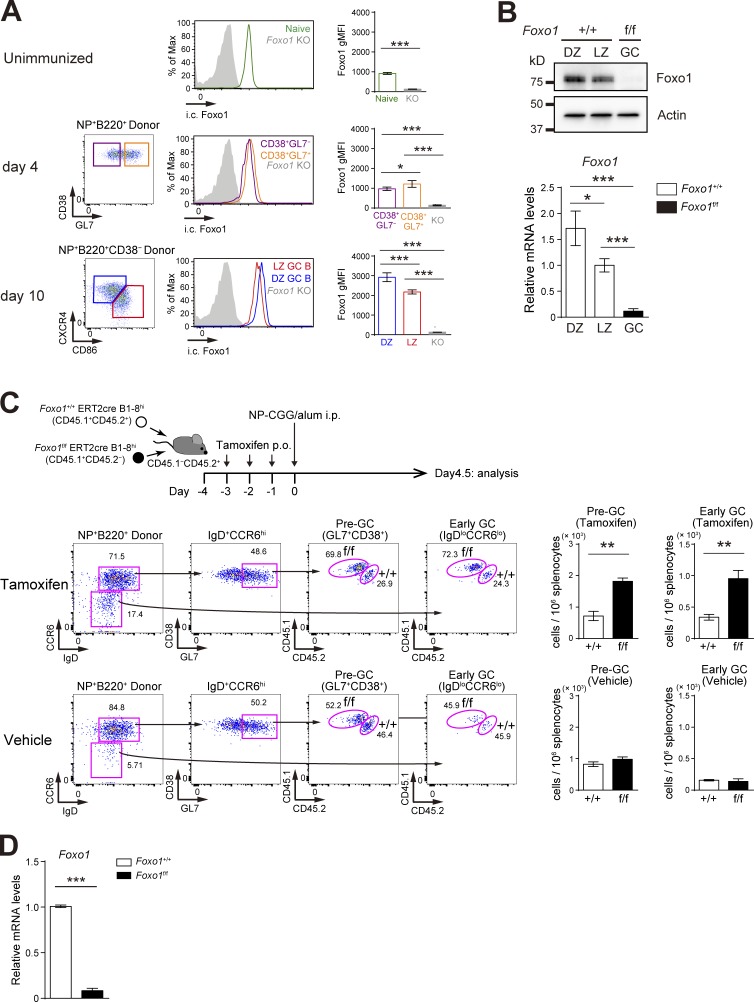
Figure 1.
Hyperexpansion of preGC B cells with Foxo1 ablation. (A) Left, flow cytometry of intracellular Foxo1 protein expression in naive B cells at day 0 (CD45.1+B220+NP+CD38+), activated B cells on day 4 (CD45.1+B220+NP+CD38+GL7− or CD45.1+B220+NP+CD38+GL7+), LZ (CD45.1+B220+NP+CD38−CD86hiCXCR4lo), and DZ (CD45.1+B220+NP+CD38−CD86loCXCR4hi) GC B cells on day 10. Wild-type mice were transferred with B1-8hi CD45.1+ B cells and then immunized i.p. with NP-CGG/alum on day 0. Foxo1 KO staining controls (gray histograms) were prepared as previously described in Figs. 1 C and 2 A. (right) Histograms indicating the geometric mean fluorescence intensity (gMFI) of each population. n = 4 biological replicates. (B) Analysis of Foxo1 protein and mRNA expression in DZ and LZ GC B cells by Western blot (top) and real-time qPCR (bottom). Foxo1-proficient LZ, DZ, and Foxo1-deficient GC B cells were sorted from mice prepared as described in Fig. 2 A. Actin, loading control. n = 3 biological replicates. (C, top) Schematic illustration of the experimental protocol. (bottom left) Flow cytometry of NP-specific donor B cells (CD45.1+B220+NP+). (bottom right) Histograms representing the cell number of preGC (Donor B220+NP+GL7+CD38+IgD+CCR6hi) and early GC (Donor B220+NP+IgDloCCR6lo) B cells in 106 splenocytes. n = 3 biological replicates. (D) Real-time qPCR analysis of Foxo1 mRNA expression in Foxo1+/+ERT2cre B1-8hi and Foxo1f/f ERT2cre B1-8hi preGC B cells. Error bars represent SD. Data are representative of three (A) or two (B and C) independent experiments, and from one experiment with three biological replicates (D). *, P < 0.05; **, P < 0.01; ***, P < 0.001; unpaired Student’s t test (A, B, and D) and paired Student’s t test (C).