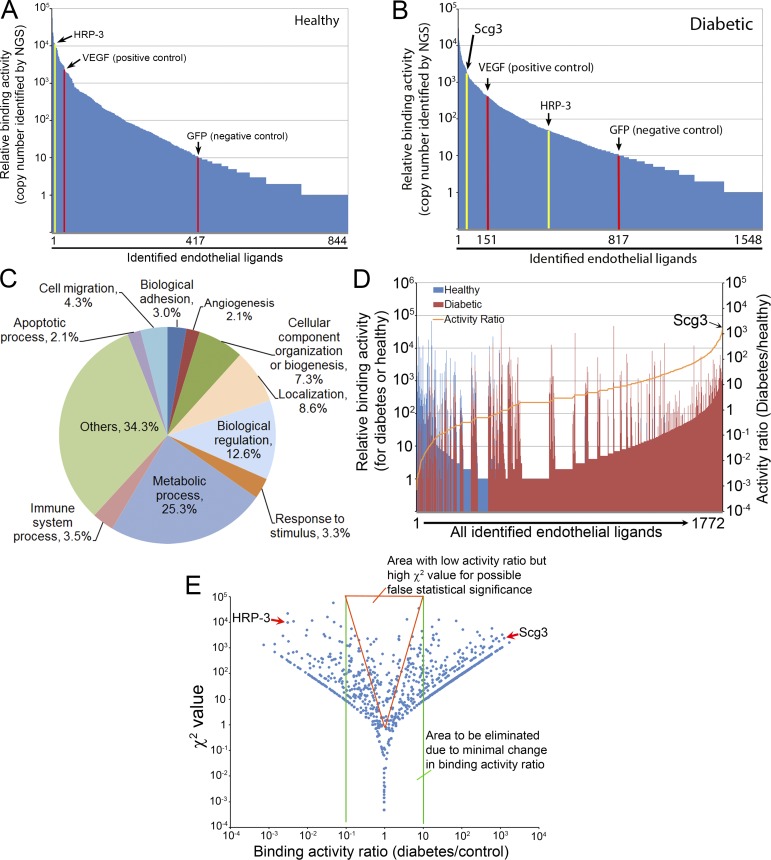
Figure 2.
Ligandomic profiling. (A and B) Ligandome profiles. A total of 844 and 1,548 putative ligands were identified by ligandomics for control (A) and diabetic (B) retina. VEGF-Phage and GFP-Phage were internal positive and negative controls, indicated by red columns. DR-high Scg3 and DR-low HRP-3 are indicated by yellow columns. Scg3 was not detected in healthy retina. GFP is used as the baseline of nonspecific binding to distinguish 417 and 817 ligands in control and diabetic retina with binding activity higher than the background. (C) Protein classification based on their roles in biological processes. (D) Activity ratio analyses to summarize the distribution patterns of binding activity (left Y axis for vertical bars) and activity ratios (right Y axis for the orange curve) for all 1,772 nonredundant ligands identified in the two conditions. Scg3 was identified with the highest binding activity ratio. (E) χ2 value distribution. The reliability of χ2 test is analyzed by plotting χ2 value against binding activity ratio. The results indicate that many ligands within the red triangle area have minimal binding activity changes but high χ2 values, suggesting that χ2 test alone may result in a large number of false positives. Based on this analysis, stringent criteria described in Results were used to identify 353 DR-high ligands and 105 DR-low ligands.