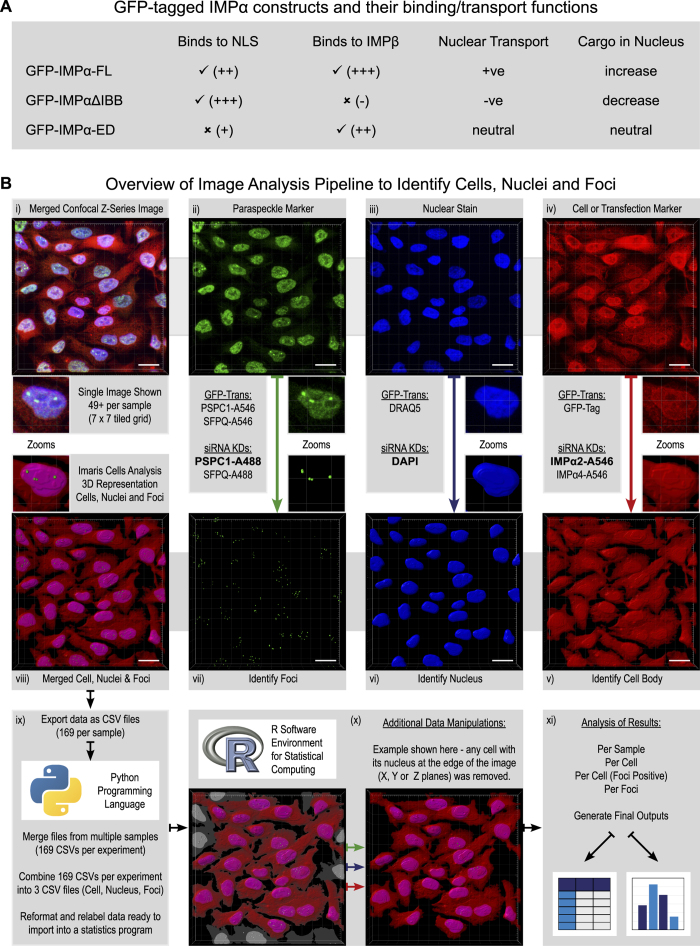
Figure 1. Overview of experimental and analytical approaches used to identify changes to subnuclear foci in response to modulating the cells nuclear transport capacity.
Using either transient transfection with plasmids encoding GFP-tagged IMPα2/α4/α6 variants or siRNA knockdown of IMPα2/α4 the capacity of IMPαs to modulate delivery of PSPC1/SFPQ into the nucleus/paraspeckles was investigated. All images are Z-series captured via confocal laser scanning microscopy, scale bars represent 20 μm. (A) GFP-tagged IMPα isoforms and their functional properties. Binding is indicated as true (✓) or false (✗), with an indication of binding strength (+/−). (B) Overview of image analysis pipeline. From this example merged z-series confocal image (Bi), the immunofluorescent signal for endogenous PSPC1 (Bii) was used to identify foci (Bvii), the nuclear marker DAPI (Biii) was used to identify the nuclei (Bvi) and the immunofluorescent signal for endogenous IMPα2 (Biv) was used to identify the cell body (Bv). The other options for paraspeckle marker, nuclear stain and cell or transfection marker for the GFP-tagged IMPα experiments (GFP-Trans) or siRNA knock down experiments (siRNA KDs) are listed. The full 3D reconstruction of cells, their nuclei and foci (Bviii) is shown for this example image but it should be noted that each sample in an experiment has 49 + of these images. Data about these cells, their nuclei and foci were exported from Imaris in CSV file formats and reorganised ready for statistical analysis using custom python programming language scripts (Bix). Additional data manipulations were performed on a per experiment basis (as detailed for each) using custom R scripts (Bx), before final statistical analysis and outputs were generated using the R software environment for Statistical Computing (Bxi). The Python logo is a trademark of the Python Software Foundation (https://www.python.org; v2.7). The R logo (https://www.r-project.org/logo/) is licensed under CC BY-SA 4.0; the license terms can be found on the following link: (https://creativecommons.org/licenses/by-sa/4.0/).