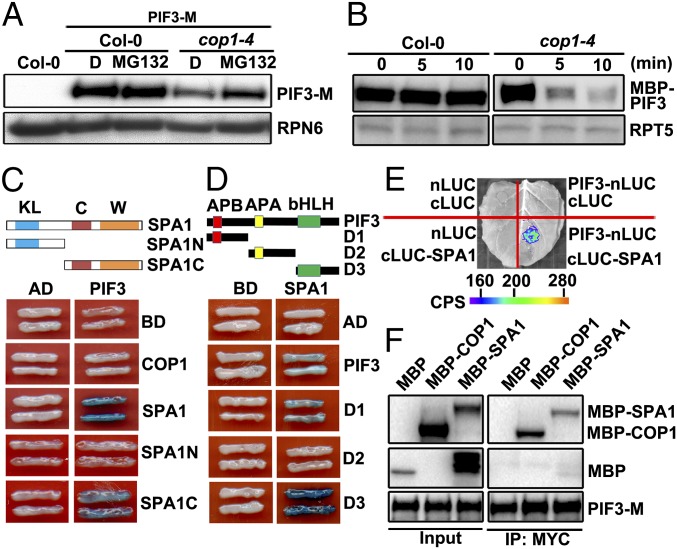
Fig. 1.
COP1/SPA complex stabilizes and interacts with PIF3 in the dark. (A) Protein levels of PIF3-M in Col-0 and cop1–4 background with or without MG132 treatment. RPN6 was used as a loading control. D, DMSO. (B) Cell-free degradation of MBP-PIF3 protein in the extracts of Col-0 and cop1–4. RPT5 was used as a loading control. (C) PIF3 interacts with SPA1 in yeast cells. C, coil domain; KL, kinase-like domain; SPA1N (residues 1–545 aa); SPA1C (residues 546–1,029 aa); W, WD40 domain. (D) Mapping of SPA1-interacting regions of PIF3 in yeast cells. Full-length and truncated PIF3 were fused with AD. Full-length SPA1 was fused with BD. PIF3-D1 (1–180 aa), D2 (181–338 aa), D3 (339–524 aa). (E) LCI assays showing the interaction of SPA1 and PIF3 in N. benthamiana leaf cells. cLUC, the vector containing C-terminal fragment of firefly luciferase; nLUC, the vector containing N-terminal fragment of firefly luciferase. Empty vectors were used as negative controls. (F) The Co-IP assays showing COP1/SPA1 complex associates with PIF3.