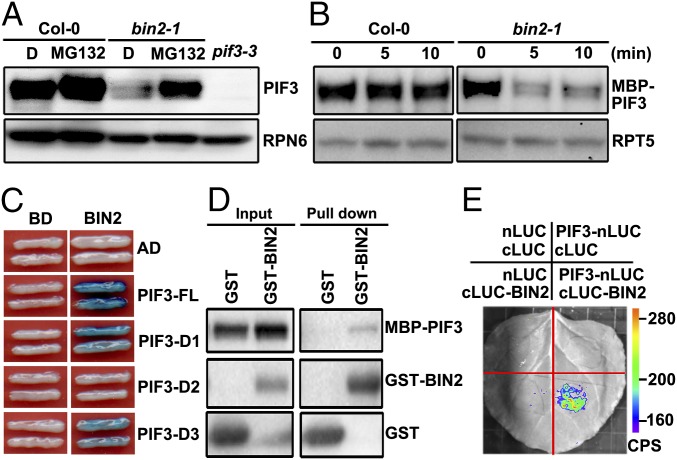
Fig. 2.
Constitutive activation of BIN2 mediates the proteasomal degradation of PIF3 in the dark. (A) Levels of PIF3 in Col-0 and bin2-1. RPN6 was used as a loading control. D, DMSO. (B) Cell-free degradation of MBP-PIF3 protein was accelerated in bin2-1 mutant extracts. RPT5 was used as a loading control. (C) BIN2 interacts with PIF3 in yeast cells. Full-length and truncated PIF3 were fused with AD. Full-length BIN2 was fused with BD. (D) In vitro pull-down assays showing the direct interaction of MBP-PIF3 with GST-BIN2. MBP-PIF3 was used as the prey molecules and incubated with GST-BIN2 or GST. (E) LCI assays showing the interaction of BIN2 with PIF3 in N. benthamiana leaf cells.