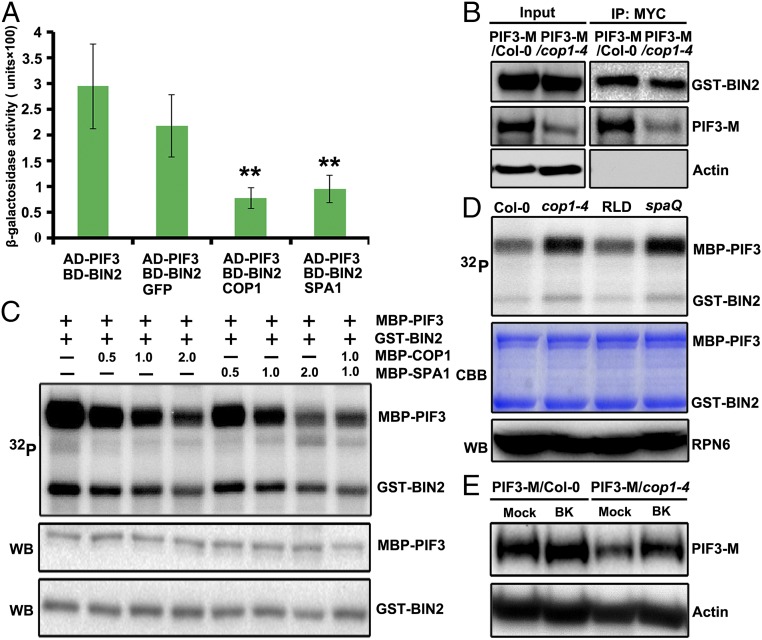
Fig. 5.
COP1/SPA complex inhibits BIN2-mediated phosphorylation and degradation of PIF3. (A) Yeast three-hybrid assays showing the COP1 and SPA1 inhibition of the BIN2–PIF3 interaction in yeast cells. Data are Mean ± SD, n = 3. **P < 0.01 (t test). (B) The Co-IP assays showing a stronger interaction between GST-BIN2 and PIF3 in the absence of COP1. Actin was used as a loading control. (C) COP1/SPA1 inhibits BIN2-mediated phosphorylation of PIF3 in vitro. The quantity (micrograms) of MBP-COP1 or MBP-SPA1 added in the reaction mix was as indicated. (D) Cell-free kinase assays showing the phosphorylation levels of the MBP-PIF3 in the total extracts of dark-grown Col-0, cop1–4, RLD, and spaQ seedlings. (Top) the autoradiogram of MBP-PIF3; (Middle) CBB staining of the gel; (Bottom) RPN6 was used as a loading control. (E) Protein gel blot of PIF3-M in Col-0 and cop1–4 treated with or without 30 μM BK. Actin was used as a loading control.