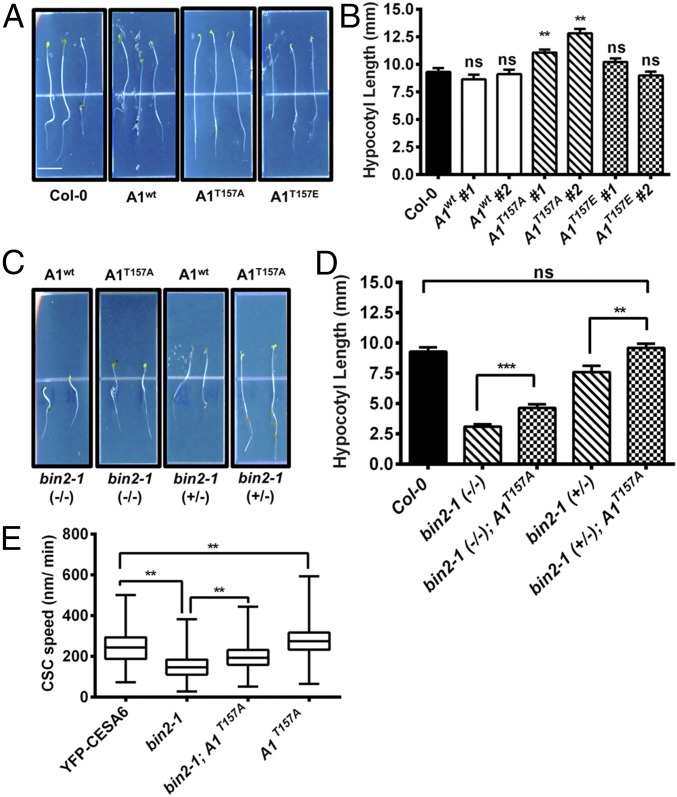
Fig. 4.
Mutations of BIN2-targeted CESA1 phosphorylation sites restore growth and CSC velocity in bin2-1. Arabidopsis cesa1-null mutants were transformed with CESA1wt or CESA1T157 phosphorylation site mutants, and the growth behavior of these mutants was examined in 5-d-old dark-grown hypocotyls compared with wild-type Col-0 controls. (A) Representative images of CESA1T157A and CESA1T157E phosphorylation site mutants compared with wild-type Col-0 seedlings or cesa1-null mutants transformed with CESA1wt. (B) Quantification of hypocotyl lengths in seedlings from A. The means and SEM are presented as in Fig. 1B. One-way ANOVA revealed a significant difference: F(6, 518) = 16.32 (**P < 0.01; n = 58–103). ns, not significant from wild-type control by Tukey post hoc analysis. The CESA1T157A mutant was introgressed into a segregating population of the bin2-1 mutant to determine if CESA1T157A could partially complement bin2-1. (C) Representative images of bin2-1 (+/−) or bin2-1 (−/−) plants with or without CESA1T157A. (D) Quantification of hypocotyl lengths from C. Error bars represent SEM (n = 19–30). One-way ANOVA revealed a significant difference: F(4, 198) = 34.26. Multiple Student’s t tests were performed against the indicated conditions (**P < 0.01, ***P < 0.005). ns, not significant. (E) CSC velocities were measured in the bin2-1 background with or without CESA1T157A and in the CESA1T157A mutant by spinning disk microscopy. These results were compared with YFP-CESA6 control CSC velocities. Error bars represent minimum and maximum values (n = 320–1,784 complexes). One-way ANOVA revealed a significant difference: F(3, 4,379) = 1057 (**P < 0.01 by Tukey post hoc analysis between the indicated conditions).