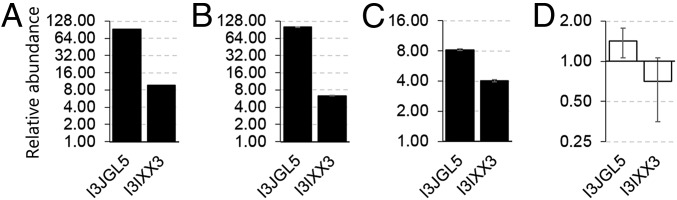
Fig. 1.
Targeted SWATH-MS/Skyline protein quantitation of IMPA1.1 and MIPS enzymes in cells grown in hyperosmotic (650 mOsmol/kg) medium relative to isosmotic (315 mOsmol/kg) controls, for which the abundance is 1. Proteomic analysis was performed on tryptic MIPS and IMPA1.1 peptides whose sequence is 100% conserved between O. mossambicus and O. niloticus using the O. niloticus proteome as a reference. Hence, accession numbers are I3JGL5 for IMPA1.1 and I3IXX3 for MIPS. Columns shown in black indicate a significant increase in protein abundance under hyperosmotic conditions (Benjamini–Hochberg adjusted P < 0.01). White columns indicate no significant effect of hyperosmolality (P > 0.05). (A and B) The increase in the abundance of IMPA1.1 and MIPS proteins after 72 h hyperosmolality did not differ in untransfected OmB cells (A) and transfected OmB cells (B). (C and D) In addition, both proteins were up-regulated after 24 h of hyperosmotic stress in the absence of actinomycin D (C), whereas the hyperosmotic up-regulation was completely inhibited by 10 µM actinomycin D (D). Data shown are means ± SEM, n = 5. Quantitative data and spectral libraries are accessible at Panorama public (https://panoramaweb.org/labkey/XW2016-1.url).