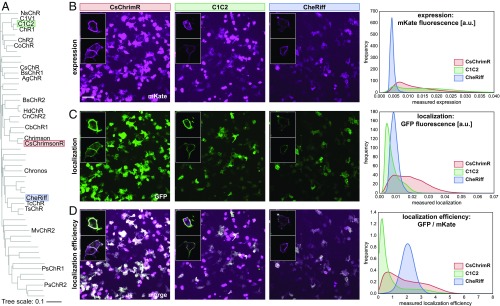
Fig. 1.
Parental ChRs and their properties. (A) Phylogenetic tree of published ChR sequences. Sequences with an alias (e.g., NsChR) have been characterized for expression and functionality in HEK cells and/or mammalian neurons. The three parental sequences (C1C2, CsChrimsonR, and CheRiff) are highlighted. (B–D) HEK cells were transfected with a parental ChR. Membrane-localized ChR was labeled using SpyCatcher-GFP assay, and ChR expression was measured using mKate. HEK cell populations were imaged and processed to measure expression [mean mKate fluorescence (in arbitrary units)], plasma membrane localization [mean GFP fluorescence (in arbitrary units)], and localization efficiency (mean GFP fluorescence/mean mKate fluorescence). Example images show population expression (B), localization (C), and localization efficiency (D) for each parental construct. For both CsChrimR and C1C2, there are cells with very high levels of mKate signal that do not perfectly colocalize with the GFP localization label. These cells express at high levels, much of which is not trafficked to the plasma membrane. (Scale bar: 100 μm.) Insets show confocal images for a few representative cells expressing each parental construct. HEK cell population images were segmented, and the ChR expression, localization, and localization efficiency were measured for each cell. The distribution of these properties for the population of transfected cells is plotted for each parent using kernel density estimation for smoothing.