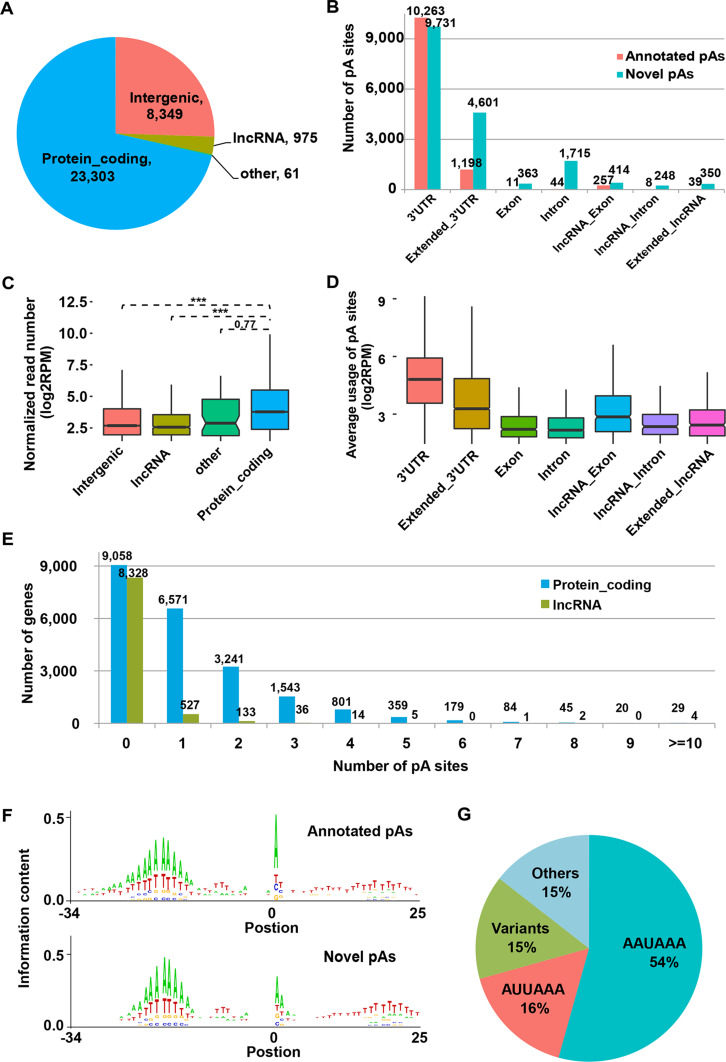
Fig. 1.
Distribution and motif sequences of the identified pA sites. a Comparison of the number of the identified pA sites located in protein-coding, long non-coding (lncRNA), other genes (pseudogenes and small non-coding genes), and intergenic regions. b Distribution of the identified pA sites across genomic regions divided into seven categories including the annotated 3′UTRs, extended 3′UTRs (Extended_3′UTR), exon (Exon) and intron (Intron) of protein-coding genes, and exon (lncRNA_Exon) and intron (lncRNA_Intron) of lncRNA genes. Number of the annotated pA sites (red) from Gencode and the novel identified pA sites (cyan) for each category was labeled. c Normalized read number (RPM) of pA sites (in logarithm scale) in the genomic regions corresponding to those defined in a (***p < 0.001). d Box plots of the average usage of the pA sites in different genomic regions for each gene. e Distribution of the number of pA sites for protein-coding (blue) and lncRNA (green) genes. f Information content of both the known (upper panel) and novel identified pA sites (lower panel) around 50 nt of the pA site. g Frequency of the polyadenylation signals (PAS), including the canonical AAUAAA and 12 most common variants, reported in human in the upstream 10–30 nt of the pA sites