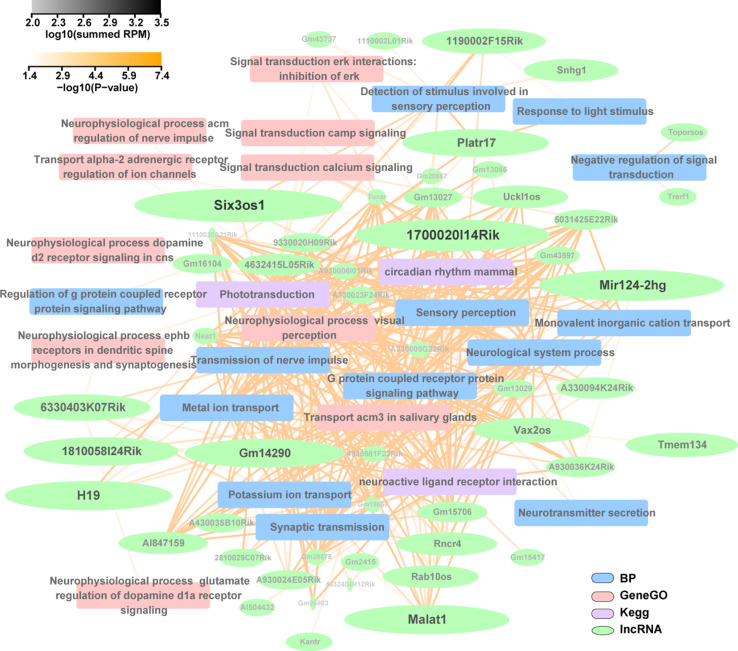
Fig. 7.
Polyadenylated lncRNA/mRNA co-expression network associated with retinal functions constructed using the Cytoscape program. The lncRNAs with a minimum expression level (log10 summed RPM) of 2 and a lncRNA-mRNA Pearson correlation coefficient larger than 0.9 were selected to draw the functional annotation network. The nodes indicate lncRNAs (green), biological processes of GO enrichment (BP, blue), KEGG (purple), and GeneGo (red) pathways. The transparency of the edges (yellow) represents the significance of the functional annotation. The text transparency and font size (black to light grey) of the lncRNA names are scaled to the estimated expression level of the corresponding lncRNAs