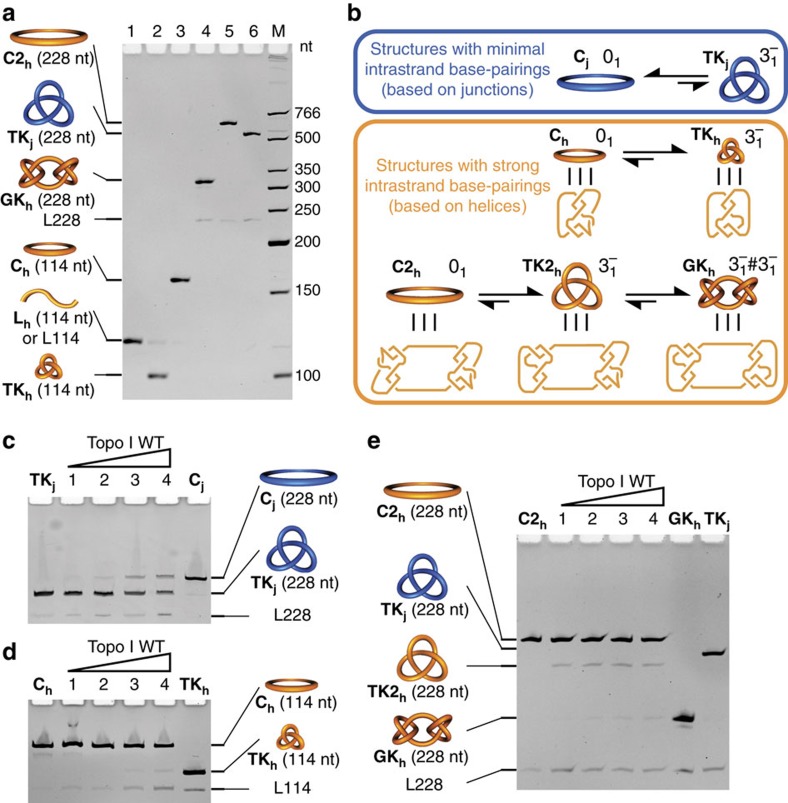
Figure 4. Comparing probes for RNA topoisomerase (RNA Topo) activity.
(a) dPAGE analyses of various structures: monomeric linear (Lh, lane 1), trefoil knot (TKh, lane 2), and circular (Ch, lane 3) RNA molecules with the sequence for helix-based topological structures; dimeric granny knot (GKh, lane 4), and circular (C2h, lane 5) species with the same sequence; and the junction-based negative trefoil knot (TKj, lane 6). (b) Hypothetical conversions of junction-based (between Cj and TKj) and helix-based (between Ch and TKh, and between C2h, TK2h and GKh) topological structures under ‘ideal' RNA Topo condition, when the RNA strand-passage event can freely take place. (c–e) Topological relaxations of TKj (c), Ch (d) and C2h (e) catalysed by increasing concentrations of WT E. coli DNA Topo I (30 min incubation at 37 °C). In c,e, the RNA probe substrates (TKj or C2h) were 80 nM and Topo I in lanes 1–4 was 40, 80, 160 and 320 nM. In d, the RNA probe substrate (Ch) was 160 nM and Topo I in lanes 1–4 was 80, 160, 320 and 640 nM. In e, C2h is relaxed to the trefoil knot TK2h and to GKh after one and two strand-passage events, respectively. All lanes contain the linear break-down products of the closed RNA structures and they are annotated by LX, in which X represents X-mer.