Abstract
The rapid and accurate quantification of the pathogenic bacteria is extremely critical to decrease the bacterial infections in all areas related to health and safety. We have developed an electrochemical strategy for simultaneous ultrasensitive detection of E. coli O157:H7 and Vibrio cholerae O1. This approach was based on the specific immune recognition of different pathogenic bacteria by multifunctional nanoconjugates and subsequent signal amplification. By employing the proposed biosensor, the concentrations of these pathogenic bacteria could be established on a single interface in a single run with improved sensitivity and accuracy. The successful approach of the simultaneous detection and quantification of two bacteria by an electrochemical biosensor demonstrated here could be readily expanded for the estimation of a variety of other pathogenic bacteria, proteins, and nucleotides. Because of their high sensitivity, electrochemical biosensors may represent a new avenue for early diagnosis of diseases.
Keywords: Electrochemical biosensor, Multifunctional nanoconjugates, Simultaneous detection, Escherichia coli O157:H7, Vibrio cholerae O1.
Introduction
Pathogenic bacteria widely exist in human habitats. As the causative agents of various infectious diseases, several bacteria, such as Escherichia coli O157:H7 (E. coli O157:H7) and Vibrio cholerae O1, if not treated properly, would often cause death 1-3. Normally, the pathogenic bacteria infect humans through the food, water, and air. To decrease bacterial infections, rapid and accurate quantification of the pathogenic bacteria is urgently needed in all health- and safety-related areas, such as clinical diagnosis, disease prevention, environmental analysis and food safety 4. Normally, the assays for pathogenic bacteria are performed based on the molecular diagnostic methods for the detection of nucleic acids in clinical and environmental samples. Although these assays showed high sensitivity and specificity, the time-consuming and laborious preparation of target DNA limit their widespread applications. Recently, various platforms have been developed to overcome these limitations, including electrochemical biosensors, surface-enhanced Raman scattering, fluorescent assays, optical sensors, and localized surface plasmon resonance 5-10.
Electrochemical biosensors, as a powerful analytical technique with simple preparation, fast analysis, high sensitivity and uncomplicated instrumentation, have attracted significant attention. Thus, it was considered as an artificial choice for efficient and sensitive detection of pathogenic bacteria 11-13. Recently, we have developed sensitive electrochemical biosensors for amperometric detection of E. coli O157:H7 and acceptable applications have been obtained based on these proposed electrochemical biosensors 14-16. Most of these techniques detect single species of pathogenic bacteria on a single interface. This approach, although very useful, does not meet the demands for clinical diagnosis, especially for early diagnosis of acute bacterial infections entailing multiple pathogenic bacterial infections with similar clinical symptoms. Although challenging, it is critical to develop multiplexed electrochemical biosensors for simultaneous detection of different pathogenic bacteria.
With the emergence of sophisticated biotechnology and nanotechnology techniques, various electrochemical strategies have been described for concurrent detection of chemicals and proteins 17-21. Lu et al demonstrated the simultaneous detection of adenosine and cocaine in a single solution with Quantum dot-encoded aptamer sensors 17. Hansen and coworkers reported a sensitive and selective bioelectronic assay for several proteins by coupling aptamers with the amplification features of inorganic nanocrystals 19. Although significant advantages have been achieved by using these strategies, improvements are still needed to improve their sensitivity and applicability for simultaneous detection of different pathogenic bacteria.
The critical challenge was to improve the detection sensitivity based on an efficient amplification strategy. The normal immune-technology methods for the detection of pathogenic bacteria do not permit the amplification of antibodies directly. Recently, nucleic acids have gained special attention for signal amplification, where various approaches with excellent design and plasticity, such as hybridization chain reaction (HCR), could be employed. Cascade amplification could be obtained via HCR by extending from an initiator (short, specific nucleotide sequence) to a long repeated nucleotide sequence 22-27. Therefore, a significant enhancement of the detection signal and sensitivity could be expected by labeling the detection antibody of pathogenic bacteria with nucleotide sequences, introducing HCR amplification, and attaching a large number of signal indicators on these repeated nucleotide sequences. Another strategy to improve the detection sensitivity was to increase the number of labeled signal indicators on signal probes. Normally, the signal probes could be conjugated to the antibodies directly. With the rapid development of nanotechnology, a variety of nanomaterials with large surface areas have been widely employed as nanocarriers to increase the number of signal probes modified on the antibodies. In this respect, gold nanoparticles (AuNPs), because of their good conductivity and biocompatibility, were used for improving the detection sensitivity. Recently, C60 has gained credence in the electrochemistry field because of its very rich electronic properties. C60 can adsorb metal nanomaterials, such as AuNPs to form AuNPs modified C60 (C60@AuNPs) via intermolecular bonds and molecule-substrate interactions. Subsequently, C60@AuNPs can modify a large number of antibodies or nucleic acids as nanocarriers for an increased sensitivity and an improved detection limit 28. Considering the potential of C60@AuNPs and high amplification efficiency of HCR, we combined the C60@AuNPs as nanocarriers to adsorb a large number of initiators and generate HCR amplification. This combination resulted in exponentially large number of signal indicators forming efficient multifunctional nanoconjugates for ultrasensitive detection of pathogenic bacteria.
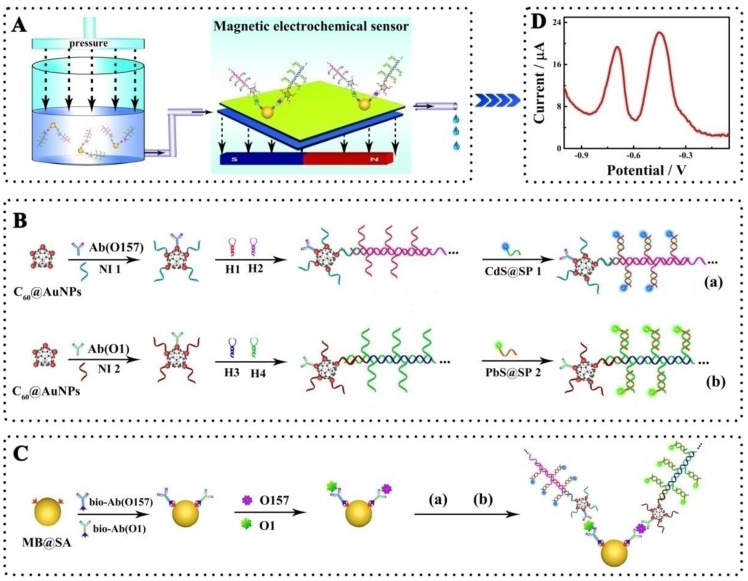
Herein, we propose a simultaneous electrochemical strategy for the ultrasensitive detection of E. coli O157:H7 and Vibrio cholerae O1 based on the specific immune recognition of these pathogenic bacteria and signal amplification via multifunctional nanoconjugates. First, the biotinylated antibodies for E. coli O157:H7 and Vibrio cholerae O1 were attached to streptavidin-coated magnetic beads (MB@SA). Next, the detection antibodies were labeled with exponentially large amounts of signal indicators (CdS and PbS nanoparticles)-labeled signal probes via C60@AuNPs as nanocarriers and HCR amplification. Thus, the biosensor could capture functionalized detection antibodies by antibody-antigen interactions in the presence of the E. coli O157:H7 and Vibrio cholerae O1. The electrochemical signals generated on different potentials were significantly enhanced and were proportional to the concentration of E. coli O157:H7 and Vibrio cholerae O1. Based upon our results presented in this study, it could be speculated that, in the future, electrochemical biosensors would be of great clinical significance. Highly sensitive and simultaneous monitoring of several pathogenic bacteria and other substances is expected to enable early detection of a variety of diseases.
2. Experimental
2.1. Reagents and Materials
Monoclonal antibodies to E. coli O157:H7 (ab20976 and ab13625) and polyclonal antibodies to Vibrio cholerae O1 (ab79794 and ab79793) were purchased from Abcam (Cambridge, MA, USA). Heat-killed E. coli O157:H7 and Vibrio cholerae O1 were a gift from the Department of Clinical Microbiology and Immunology, College of Laboratory Medicine, Third Military Medical University (Chongqing, China). MB@SA, C60, mercaptoacetic acid (MPA), 1-ethyl-3-[3-(dimethylamino)propyl] carbodiimide hydrochloride (EDC), gold chloride (HAuCl4) and sodium citrate were purchased from Sigma Chemical Co. (St. Louis, MO, USA). All other materials used were of analytical grade and purchased from Chemical Reagent Company (Chongqing, China). Phosphate buffered saline (PBS) was prepared with Na2HPO4, KH2PO4, and KCl, and PBS with tween-20 (PBST) was prepared by adding 10% tween-20 to PBS. Ultrapure water with a resistivity of 18.2 MΩ/cm was used for all reagents.
All oligonucleotides were custom-synthesized by Shanghai Sangon Biological Engineering Technology and Services Co., Ltd. (Shanghai, China). Details of the oligonucleotides are listed in Table S1. Before using, all of the hairpin nucleotides were heated to 95 °C for 2 min and then cooled to room temperature to form stem-loop structures.
2.2. Apparatus
Electrochemical measurements including cyclic voltammetry (CV), anodic stripping voltammetry (ASV) and electrochemical impedance spectroscopy (EIS) measurements were performed by a Zennium electrochemistry workstation (ZAHNER-Elektrik GmbH & Co. KG, Germany) with a conventional three-electrode system containing a platinum wire as auxiliary electrode, a saturated calomel electrode (SCE) as reference electrode and a glassy carbon plate (GCP) with/without magnetic adsorption of magnetic beads as working electrode, respectively. The morphological characterizations were carried out with JSM-7800 Extreme-resolution analytical field emission scanning electron microscope (SEM, Jeol Inc., MA, USA) and Tecnai G2 F20 field-emission high-resolution transmission electron microscope (STEM, FEI, USA). Raman spectrometer was obtained using a Renishaw Invia Raman spectrometer with 50-objective, which was calibrated first by a silicon wafer at 520 cm-1 Raman shift before each measurement.
2.3. Preparation of the Biotinylated Antibodies
The antibodies for E. coli O157:H7 and Vibrio cholerae O1 were biotinylated according to the manufacturer's instructions 29. Briefly, the appropriate NHS-biotin solution was added into the PBS solution with antibodies for E. coli O157:H7 (1 mg/mL) for the reaction between NHS group on NHS-biotin and amine groups on antibodies. The reaction was carried out at 37 °C for 2 h after mixing thoroughly. Finally, the biotinylated antibodies for E. coli O157:H7 were purified by desalination. The biotinylated antibodies for Vibrio cholerae O1 were prepared based on a similar protocol. The biotinylated antibodies were stored at 4 °C until further use.
2.4. Preparation of PbS and CdS Nanoparticles
PbS nanoparticles were prepared according to the literature with MPA as a stabilizer 30. Briefly, the Pb(NO3)2 and Na2S solutions were filtered through a 22 μm microporous membrane filter. Subsequently, 9.22 μL MPA was added to 50 mL 0.4 mM Pb(NO3)2 solution. After adjusting pH to 7 and bubbling with nitrogen for 30 min, 15 mL Na2S solution (1.34 mM) was slowly added to the mixture solution. Under nitrogen protection, the reaction was carried out for 24 h to obtain PbS nanoparticles.
CdS nanoparticles were prepared according to the previously described method with minor modifications 31. Briefly, 90 μL MPA was added into 20 mL 20 mM CdCl2 solution and the pH was adjusted to 10 with 1 M sodium hydroxide solution. Subsequently, 20 mL 20 mM thioacetamide was slowly added to the solution with continuous stirring. The mixture was refluxed at 80 °C for 10 h to obtain CdS nanoparticles. After purification, the CdS nanoparticles were re-dispersed in ultrapure water and kept at 4 °C when not in use.
2.5. Preparation PbS and CdS Nanoparticles-Functionalized Oligonucleotide
The PbS nanoparticles were labeled on to oligonucleotides according to previous literature with a minor modification 32. Briefly, 15 μL 0.1 M imidazole solution (pH 6.8) was added to 100 μL 5-amino group-capped oligonucleotides (signal probe 1, 100 μM) with complete mixing. Next, the mixture and 25 μL 0.1 M EDC were added to 2.0 mL PbS nanoparticles solution with magnetic stirring, and the reaction was kept at room temperature for 24 h with continuous stirring. The synthesized oligonucleotide bioconjugated PbS nanoparticles (PbS-SP) were collected by centrifugation at 11000 rpm and re-suspended in water. The PbS-SP were stored at 4 °C. The oligonucleotide bioconjugated CdS nanoparticles (CdS-SP) were prepared by the same protocol.
2.6. Preparation of Functionalized Detection Antibodies
As shown in Scheme 1, the antibodies were multi-functionalized with oligonucleotide bioconjugated CdS or PbS nanoparticles and amplified with HCR amplification. Typically, the C60 nanoparticles were first modified with thiol groups by the condensation of 30 mg C60 powder, 18.5 mg sarcosine, and 8.3 mg 10-mercaptodecyl aldehyde in 90 mL toluene and refluxing for 24 h under the N2 atmosphere 33. After purification, the obtained thiolated C60 nanoparticles were modified with AuNPs, which were prepared according to the literature and characterized with average particle size of 16 nm 34. In a typical experiment, 300 μL of prepared AuNPs were added into 1.7 mL thiolated C60 solution and the mixture was held with magnetic stirring at 4 °C for 12 h. After purification, the AuNPs-modified C60 nanoparticles (C60@AuNPs) were stored at 4 °C for further use.
Scheme 1.
Schematic representation of the simultaneous detection of pathogenic bacteria by the proposed electrochemical biosensor using E. coli O157:H7 and Vibrio cholerae O1 as a model: A, Schematic diagram of the simultaneous electrochemical system; B, Preparation of the bioconjugates- and nanoparticles-labeled antibodies; C, Schematic diagram of the immunoassay on magnetic beads. Abbreviations: Ab(O157) or Ab(O1), antibodies for E. coli O157:H7 or Vibrio cholerae O1; Bio-Ab, biotinylated antibodies; H1-4, hairpin nucleotide for HCR reaction; MB@SA, streptavidin-coated magnetic beads; NI1-2, nucleotide initiator; SP1-2, signal probe. (The schematic diagram is helpful for understanding the fabrication of the electrochemical assay, but it is not in proportion).
To conjugate the signal probes (CdS- or PbS-modified oligonucleotides) on antibodies and amplify with HCR amplification, 10 μL nucleotide initiator 1 and 2 μL detection antibodies for E. coli O157:H7 were added to 1 mL C60@AuNPs solution and the mixture was kept at 4 °C for 12 h to functionalize the C60@AuNPs. 100 μL 0.1 M HT solution was then added to the mixture to block the nonspecific sites. The prepared nucleotide initiator-labeled antibodies with AuNPs@C60 was incubated with 1 μM hairpin nucleotide 1 and 2 in 6.67 mM Tris-HCl buffer (pH 8.0) containing 66.67 mM MgCl2 for 30 min. The resultant product, detection antibodies for E. coli O157:H7 labeled with a large amount of repeated nucleotide sequence based on HCR reactions, was then further reacted with 6 mM PbS-labeled signal probe (PbS-SP) by hybridization between the signal probe and associated sequences on the long repeated nucleotide sequences. The nucleotide initiator 2 and detection antibodies for Vibrio cholerae O1 was prepared and further connected with the CdS-labeled signal probe (CdS-SP) using the same protocol.
2.7. Measurement Procedure
Before the electrochemical measurements, the glassy carbon plate (GCP) was polished with alumina slurries (0.3 and 0.05 µm, respectively), ultrasonicated in ethanol and distilled water, and dried with nitrogen. Subsequently, the cleaned GCP was modified with a gold nano-layer by electrodeposition in HAuCl4 (1%) solution at the potential of -0.2 V for 30 s. Finally, the gold nano-layer modified glassy carbon plate (DepAu/GCP) was dipped in 0.1 M HT to block the nonspecific sites.
In this study, the electrochemical measurements were performed based on a sandwich type assay in a tube as shown in Scheme 1. Briefly, the biotinylated primary antibodies for E. coli O157:H7 and for Vibrio cholerae O1, PbS-labeled signal probe (CdS-SP)-functionalized detection antibodies for E. coli O157:H7 and CdS-labeled signal probe (CdS-SP)-functionalized detection antibodies for Vibrio cholerae O1 were first added to the solution with streptavidin-coated magnetic beads (MB@SA). After addition of test sample, the mixture was incubated at 37 °C for 40 min. Next, the mixture was employed for the electrochemical measurements with the self-designed magnetic, electrochemical sensor. After each determination, the magnetic electrochemical sensor could be regenerated simply by washing twice with PBST. Details of the magnetic, electrochemical sensor are described in the Supporting Information Figure S2.
2.8. Preparation of the Synthetic Samples and Safety Consideration
The bacteria were maintained in brain-heart infusion at 37 °C for 20 h following which the concentration was determined by the conventional PCR technology. The bacteria were heated at 100 °C for 15 min to kill all of the bacteria. The pure cultures were serially diluted to different concentration with PBS as the synthetic samples.
3. Results and Discussion
3.1. Morphology and Structure Characterization of the Nano-materials
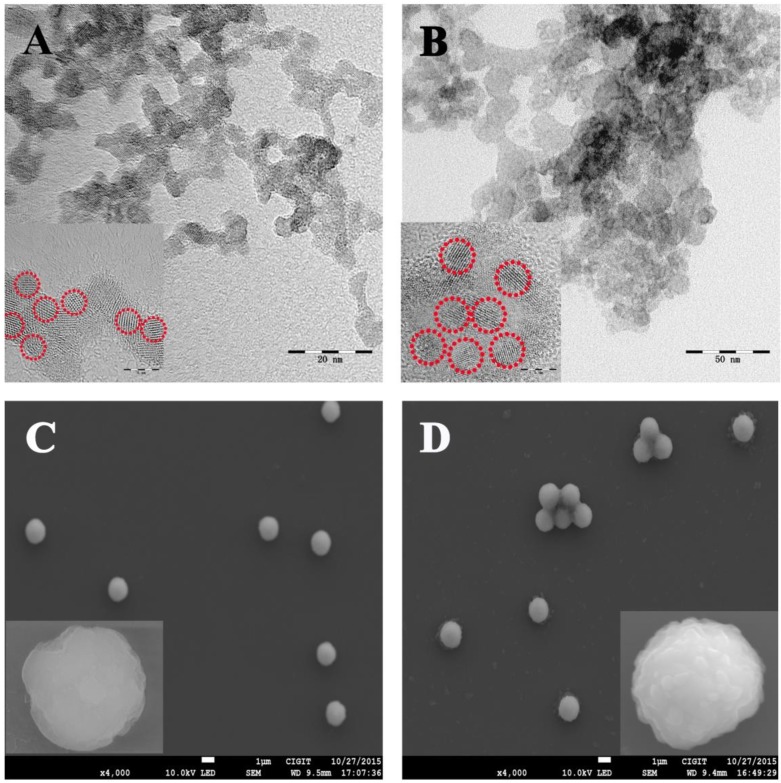
The morphology of the CdS and PbS nanoparticles were characterized by STEM (Figure 1A and B, respectively). As shown in Figure 1A, the CdS nanoparticles presented a spherical structure with an average diameter of 4 nm. For the PbS nanoparticles (Figure 1B), the average diameter was about 10 nm. The excellent nucleation and dispersion of CdS and PbS nanoparticles indicated the suitability of their application as signal probes. The MB@SA used in this study and the immunoreaction on the magnetic beads were characterized by SEM and zeta potential measurements. As displayed in Figure 1C, the SEM image of MB@SA revealed a spherical structure with an average diameter of 1.30 μm (the particle size distribution is shown in the Supporting Information Figure S5A) and zeta potential of 14.7 mV. When the capture antibodies were conjugated onto the surface of MB@SA (Figure 1D), the diameter of the beads was increased to 1.33 μm with some irregular bulges on the surface. At the same time, the zeta potential was changed to 23.2 mV due to the addition of antibodies on the surface of MB@SA Supporting Information Figure S5B). All these results indicated successful attachment of capture antibodies on the MB@SA surface which was critical for the immunoassay based on electrochemical strategy in this study.
Figure 1.
Structure and morphology of the nanoparticles: (A) CdS nanoparticles; (B) PbS nanoparticles; (C) MB@SA; (D) MB@SA/capture antibodies. Inserts show the HTEM and SEM images with high amplification for CdS (A), PbS (B), MB@SA (C) and MB@SA/capture antibodies (D).
3.2. Characteristics of the Electrochemical Strategy
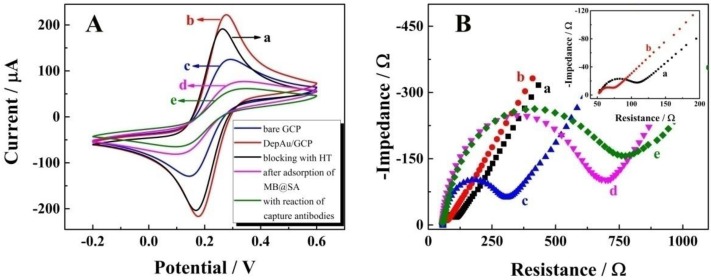
CV, the powerful electrochemical technology to characterize the information on reagent immobilization, electroactivity, and stability, was employed to study the immobilization of magnetic beads on the self-designed magnetic electrochemical sensor. As shown in Figure 2A, with the CV performance in 2 mM Fe(CN)63-/4- with 0.1 M KCl, a pair of well-defined redox peaks of Fe(CN)63-/4- could be observed for the bare GCP (curve a). After the electrochemical deposition of DepAu onto the GCP (curve b), the peak currents were increased due to the good conductivity and electron transfer ability of DepAu. When the DepAu-modified GCP (DepAu/GCP) was blocked with HT (curve c), the peak currents were decreased. After magnetic adsorption of the MB@SA (curve d), the peak currents were decreased even more indicating the stable immobilization of MB@SA which was helpful for the efficient electrochemical determination. For the electrochemical response after magnetic adsorption of the MB@SA with capture antibodies, a decreased peak current could clearly be observed (curve e). This is because the antibodies on the surface of MB@SA retard the electron transfer due to the increased steric hindrance effect.
Figure 2.
Characterization of the electrochemical measurements by self-designed magnetic electrochemical sensor: (A) CV; (B) EIS. The labeling a through e denote: a, bare GCP; b, DepAu-modified GCP; c, DepAu-modified GCP blocked with HT; d, fabricated GCP after adsorption with MB@SA; e, fabricated GCP with capture antibodies on the surface of MB@SA. Insert of Figure 2B is the EIS characterization of bare GCP (a) and DepAu modified GCP (b) with high amplification.
Further characterizations via EIS in 0.1 M PBS with 5 mM [Fe(CN)6]3-/4- at a frequency range from 5 × 10-2 to 1 × 106 Hz in a given open circuit voltage with an amplitude of 10 mV are shown in Figure 1B. The electron transfer resistance (Ret) was employed to demonstrate the electron transfer kinetics quantitatively which was similar to the semicircle diameter in EIS response in the form of a Nyquist plot. For the bare GCP (curve a), a normal EIS response was received with Ret of about 60 Ω. After the electrochemical deposition of DepAu onto the GCP surface, an obviously decreased Ret was observed (curve b, Ret ≈ 20 Ω). When the DepAu-modified GCP was blocked with HT (curve c), an increased Ret about of 200 Ω was obtained. Furthermore, the Ret was increased to about 700 Ω after magnetic adsorption of the reacted MB@SA (curve d). After magnetic adsorption of the MB@SA reacted with capture antibodies, the Ret was increased to 800 Ω due to the retardation of electron transfer via the increased steric hindrance effect (curve e). These EIS results indicated the same tendency of electron transfer as that obtained by CVs, and quantified the electron transfer kinetics with accurate Ret. These EIS and CV measurements could confirm the successful fabrication of the proposed electrochemical strategy.
3.3. Characterization of the Signal Amplification
In this study, the signal was amplified by C60@AuNPs as nanocarriers and HCR amplification. To evaluate the efficiency of signal amplification, three analysis modes were prepared for the detection of E. coli O157:H7 and Vibrio cholerae O1 simultaneously: CdS- or PbS-labeled antibodies without C60@AuNPs as nanocarriers and HCR amplification (Figure S6A), CdS- or PbS-labeled antibodies with AuNPs as nanocarriers and HCR amplification (Figure S6B), and CdS- or PbS-labeled antibodies with C60@AuNPs as nanocarriers and HCR amplification (Figure S6B). As shown in Figure S6A, there were two current peaks for PbS and CdS nanoparticles with current signals of about 5 μA. For CdS- or PbS-labeled antibodies with AuNPs as nanocarrier and HCR amplification (Figure S6B), an improvement to the electrochemical response was obtained with current signal of about 12 μA. For CdS- or PbS-labeled antibodies with HCR amplification (Figure S6C), a 2-fold enhancement was obtained. From these results, it became obvious that the amplification of C60@AuNPs as nanocarriers and HCR amplification could be employed for the detection of E. coli O157:H7 and Vibrio cholerae O1 simultaneously.
The nucleotides employed for the HCR amplification were characterized by polyacrylamide gel electrophoresis (Figure S1. The molecular weights of the nucleotides in the experiment were characterized by distribution places of the UV band from notches. The HCR reaction could be triggered by nucleotide initiator with hairpin nucleotides forming long nucleotides polymers (lane 5 and lane e). Importantly, the long nucleotides polymers with large molecular weights could attach massive signal probes for the enhancement of electrochemical responses associated with the nucleotide initiator. Thus, the electrochemical responses could be employed to indicate the concentration of nucleotide initiator and associated antibodies with improved sensitivity and efficiency.
3.4. Specific Reaction of the Electrochemical Assay
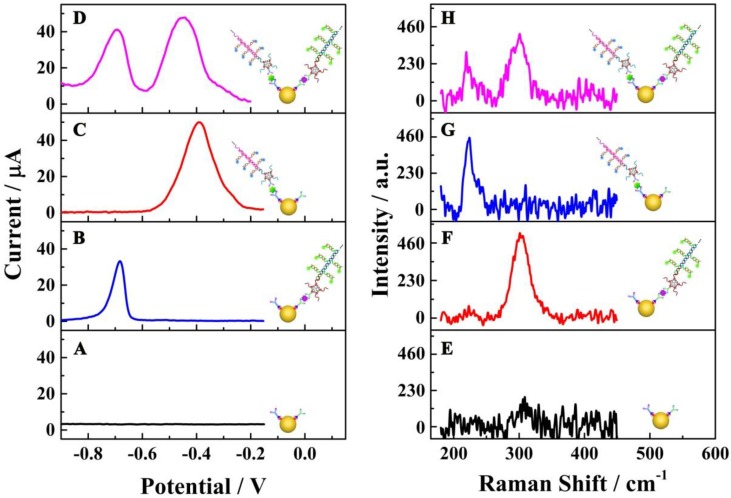
To investigate the specific reaction of the proposed multiplexed assay via electrochemical characterization, the biosensor was used to analyze different samples with the same assay mode, including 1) no targets, 2) E. coli O157:H7, 3) Vibrio cholerae O1 and 4) the mixture of E. coli O157:H7 and Vibrio cholerae O1. As shown in Figure 3A, without a target, no obvious current peak could be observed. When using E. coli O157:H7 or Vibrio cholerae alone as target, current peaks for CdS and PbS could be detected at about - 0.7 V and - 0.4 V, respectively (Figures 3B & C). When the E. coli O157:H7 and Vibrio cholerae O1 together were used as detection targets, two strong current peaks at - 0.7 V and - 0.4 V could be observed (Figure 3D). More importantly, the current peaks did not interfere with each other, indicating the signals of E. coli O157:H7 and Vibrio cholerae O1probes could be employed for their simultaneous detection on a single interface in a single run.
Figure 3.
Specific reactions of the multiplexed assay: (A-D) electrochemical characterizations; (E-H) Raman scattering. The electrochemical and Raman responses show the raw results subtracted from the background responses.
Another important technology, Raman spectrometry, was also employed to confirm the specific reactions of the electrochemical assay (Figure 3E-H). There was no obvious Raman peak from 180 cm-1 to 450 cm-1 for the interface without E. coli O157:H7 and Vibrio cholerae O1 (Figure 3E). With E. coli O157:H7 alone, there was a single Raman peak at about 300 cm-1 (Figure 3F), which was the characteristic Raman peak for CdS 35. For the detection of Vibrio cholerae O1 alone, a single Raman peak could be observed at about 230 cm-1 (Figure 3G), indicating the presence of the PbS nanoparticles 36. When the proposed biosensor was employed for the simultaneous detection of E. coli O157:H7 and Vibrio cholerae O1 on a single interface (Figure 3H), two Raman peaks were present at about 230 cm-1 and 300 cm-1 pertaining to PbS and CdS nanoparticles, respectively. This again indicated the acceptable specificity of the electrochemical strategy for the simultaneous detection of E. coli O157:H7 and Vibrio cholerae O1.
3.5. Analytical Characteristics of the Biosensor
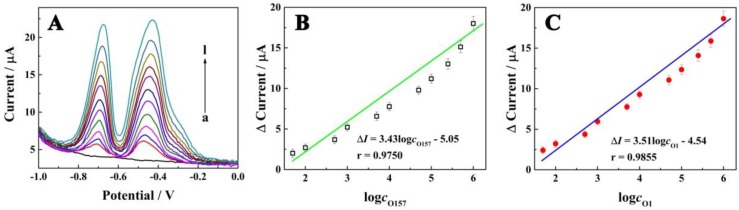
We evaluated the performance of the electrochemical biosensor for simultaneous determination of different concentrations of E. coli O157:H7 and Vibrio cholerae O1 in standard solution. As displayed in Figure 4A, the current signals were increased according to the concentrations of E. coli O157:H7 and Vibrio cholerae O1 in the range from 5×101 to 1×106 CFU/mL. Also, a good linear relationship with the logarithm concentrations of E. coli O157:H7 and Vibrio cholerae O1 can be seen in Figure 4B and C, with egression equation of ∆I (µA) = 3.43log cO157:H7 - 5.05 and ∆I (µA) = 3.51logcO1 - 4.54, and detection limit of 39 CFU/mL and 32 CFU/mL, respectively; this equation was defined as LOD = 3SB/m, where SB was the standard deviation of the blank and m was the analytical sensitivity. These results indicated that the E. coli O157:H7 and Vibrio cholerae O1 concentrations could be quantitatively measured by the electrochemical signal using the proposed biosensor. Furthermore, the performance of the proposed biosensor was compared with that of the previously reported studies as shown in Table 1. The results indicated the superior sensitivity of the biosensor for the detection of bacteria with a low detection limit and wide linear range.
Figure 4.
Electrochemical signals as a function of concentrations of E. coli O157:H7 and Vibrio cholerae. (A) Electrochemical responses of the electrochemical biosensor at different concentrations of E. coli O157:H7 and Vibrio cholerae O1 (curves a-i, with concentration of 0, 5 × 10, 1 × 102, 5 × 102, 1 × 103, 5 × 103, 1 × 104, 5 × 104, 1 × 105, 2.5 × 105, 5 × 105 and 1 × 106 CFU/mL, respectively); (B) Calibration curve for the relationship between electrochemical signals and concentrations of E. coli O157:H7; (C) Calibration curve for the relationship between electrochemical signals and concentrations of Vibrio cholerae O1.
Table 1.
Comparison between the electrochemical assay with multifunctional conjugates and other recent studies
| Method | Detection limit CFU mL-1 |
Linear range CFU mL-1 |
Reference | |
|---|---|---|---|---|
| E. coli O157:H7 | ECAa | 412 | 4.12 × 102 - 4.12 × 105 | 14 |
| FAb | 25 | 4 × 10 - 4 × 105 | 37 | |
| CLAc | 7.6 × 103 | 0 - 5 × 105 | 38 | |
| LAPSd | 2.5 × 104 | n/a | 39 | |
| PCRe | 1.3 × 104 | n/a | 40 | |
| ECA | 39 | 5 × 10 - 1 × 106 | Present work | |
| Vibrio cholerae O1 | ECA | 100 | 1.0 × 102 - 1.0 × 107 | 8 |
| FA | 50 | n/a | 9 | |
| DFMf | 1.0 × 103 | 1.0 × 103 - 1.0 × 106 | 41 | |
| ECA | 32 | 5 × 10 - 1 × 106 | Present work |
a ECA, electrochemical assay;
b FA, fluorescent assay;
c CLA, chemiluminescent assay;
d LAPS, light-addressable potentiometric sensor;
e PCR, polymerase chain reaction;
f DFM, dynamic force microscopy.
The reproducibility of the biosensor, which constitutes an important feature, was investigated by intra-assay and inter-assay coefficients of variation with 10 biosensors incubated individually with standard solutions of E. coli O157:H7 and Vibrio cholerae O1 Supporting Information Figure S7. The close current signals with intra-assay coefficient of variation 6.8% and 6.7% and inter-assay coefficient of variation 5.3% and 6.6% for E. coli O157:H7 and Vibrio cholerae O1 respectively, indicated the acceptable reproducibility of the biosensor for simultaneous detection of E. coli O157:H7 and Vibrio cholerae O1. The stability, another important factor of the biosensor, was investigated by long-term storage assay. After a storage period of 30 days at 4 ºC, the current response retained 93.6% of its initial response, suggesting the satisfactory stability of the biosensor.
3.6. Analysis of Synthetic Samples
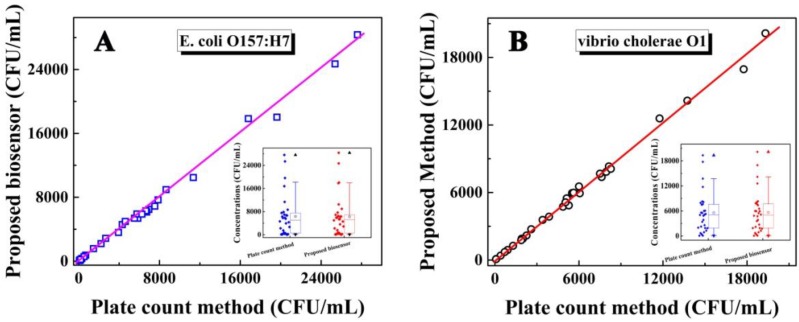
To investigate the feasibility of the biosensor for evaluating the laboratory and clinical samples, the study results of human stool samples and water samples were compared with that obtained by the plate count method. Preparation of these samples is shown in the Supporting Information. As shown in Figure 5 for the determination of E. coli O157:H7 and Vibrio cholerae O1, both the correlation coefficient and slope were close to 1, indicating a good agreement between the study results of the biosensor and those from the plate count method (insert: the concentration range of E. coli O157:H7 and Vibrio cholerae O1 obtained by the electrochemical biosensor and the plate count method, respectively). These results indicated that the biosensor was feasible for the determination of E. coli O157:H7 and Vibrio cholerae O1, could satisfy the need for practical analyses, and provided an efficient approach for the simultaneous detection of bacteria for clinical diagnosis, disease prevention, environmental analysis, and food safety.
Figure 5.
Relationship between the concentrations of E. coli O157: H7 (A) and Vibrio cholerae O1 (B) detected by the as-proposed biosensor and plate count method. Insert: The concentration range for the concentration of E. coli O157:H7 (a) and Vibrio cholerae O1 (b) respectively by box chart (box, 25%-75% value range; line, 10%-90% value range; ☆, mean value; ▲, max value; ▼, min value).
4. Conclusion
In summary, an electrochemical biosensor was developed for sensitive and efficient simultaneous determination of E. coli O157: H7 and Vibrio cholerae O1 by utilizing nanotechnology and HCR amplification with self-designed magnetic electrochemical sensor. Using the proposed biosensor, the concentrations of these pathogenic bacteria could be established on a single interface in a single run. Furthermore, the biosensor displayed excellent performance with high sensitivity with a detection limit of 39 CFU/mL and 32 CFU/mL, respectively, for E. coli O157: H7 and Vibrio cholerae O1. This high sensitivity is an essential requirement for early clinical diagnosis, disease prevention, environmental analysis, and food safety. We believe that the successful establishment of the electrochemical biosensor would provide an efficient amplification approach for ultrasensitive molecular diagnostics for the detection of multiple pathogenic bacteria on a single interface in a single run. This simple method could be expanded readily for detecting other pathogenic bacteria and would be of great value for future applications in public and environmental safety.
Supplementary Material
Supplementary figures and tables.
Acknowledgments
Financial support for this work was provided by the National Natural Science Foundation of China (No. 81401722 and 81572078).
Abbreviations
- ASV
anodic stripping voltammetry
- CdS-SP
oligonucleotide bioconjugated CdS nanoparticles or CdS labeled signal probe
- CV
cyclic voltammetry
- DepAu/GCP
gold nano-layer modified glassy carbon plate
- EDC
1-ethyl-3-[3-(dimethylamino)propyl] carbodiimide hydrochloride
- EIS
electrochemical impedance spectroscopy
- E. coli O157
H7: Escherichia coli O157:H7
- GCP
glassy carbon plate
- HCR
hybridization chain reaction
- C60@AuNPs
AuNPs modified C60
- MB@SA
streptavidin coated magnetic beads
- MPA
mercaptoacetic acid
- HAuCl4
gold chloride
- PBS
Phosphate buffered saline
- PbS-SP
oligonucleotide bioconjugated PbS nanoparticles or PbS labeled signal probe
- PBST
phosphate buffered saline with tween-20
- SCE
saturated calomel electrode.
References
- 1.Luanne H, William C, Paul S. Bacterial biofilms: from the Natural environment to infectious diseases. Nature Reviews Microbiology. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 2.Stephen S. Andrew T, ed. Plagues and Politics, Basingstoke, England: Palgrave Macmillan UK; 2001. Factors in the emergence of infectious diseases; pp. 8–26. [Google Scholar]
- 3.Tauxe R. Emerging foodborne diseases: an evolving public health challenge. Emerging Infectious Diseases. 1997;3(4):425–34. doi: 10.3201/eid0304.970403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. Foodborne illness acquired in the United States-major pathogens. Emerging Infectious Diseases. 2011;17(1):7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gustavo Z, Jordi R, Ali D, Xavier R. Immediate detection of living bacteria at ultralow concentrations using a carbon nanotube based potentiometric aptasensor. Angewandte Chemie (International ed. in English) 2009;48:7334–7. doi: 10.1002/anie.200902090. [DOI] [PubMed] [Google Scholar]
- 6.Patel S, Premasiri W, Moir D, Ziegler L. Barcoding bacterial cells: a SERS-based methodology for pathogen identification. Journal of Raman Spectroscopy. 2008;39:1660–72. doi: 10.1002/jrs.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee HJ, Kim BC, Kim KW, Kim KY, Kim J, Oh MK. A sensitive method to detect Escherichia coli based on immunomagnetic separation and real-time PCR amplification of aptamers. Biosensors and Bioelectronics. 2009;24:3550–5. doi: 10.1016/j.bios.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 8.Tam PD, Hoang NL, Lan H, Vuong PH, Anh TTN, Huy TQ, Thuy NT. Detection of vibrio cholerae O1 by using cerium oxide nanowires-based immunosensor with different antibody immobilization methods. Journal of the Korean Physical Society. 2016;68(10):1235–45. [Google Scholar]
- 9.Zamani P, Sajedi RH, Hosseinkhani S, Zeinoddini M. Hybridoma as a specific, sensitive, and ready to use sensing element: a rapid fluorescence assay for detection of Vibrio cholerae O1. Analytical & Bioanalytical Chemistry. 2016;408(23):6443–51. doi: 10.1007/s00216-016-9762-y. [DOI] [PubMed] [Google Scholar]
- 10.Wandermur G, Rodrigues D, Allil R, Queiroz V, Peixoto R, Wemeck M, Miguel M. Plastic optical fiber-based biosensor platform for rapid cell detection. Biosensors and Bioelectronics. 2014;54:661–6. doi: 10.1016/j.bios.2013.11.030. [DOI] [PubMed] [Google Scholar]
- 11.Zhao G, Zhan X. Facile preparation of disposable immunosensor for Shigella flexneri based on multi-wall carbon nanotubes/chitosan composite. Electrochimica Acta. 2010;55:2466–71. [Google Scholar]
- 12.Zhuo Y, Yuan PX, Yuan R, Chai YQ, Hong CL. Bienzyme functionalized three-layer composite magnetic nanoparticles for electrochemical immunosensors. Biomaterials. 2009;30:2284–90. doi: 10.1016/j.biomaterials.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Wang SP, Wu ZS, Qu FL, Zhang SB, Shen GL, Yu RQ. A novel electrochemical immunosensor based on ordered Au nano-prickle clusters. Biosensors and Bioelectronics. 2008;24:1020–6. doi: 10.1016/j.bios.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Cheng P, Gong JH, Fang LC, Deng J, Liang WB, Zheng JS. Amperometric immunosensor for the detection of Escherichia coli O157:H7 in food specimens. Analytical Biochemistry. 2012;421:227–33. doi: 10.1016/j.ab.2011.10.049. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Fang LC, Cheng P, Deng J, Jiang LL, Huang H, Zheng JS. An electrochemical immunosensor for sensitive detection of Escherichia coli O157:H7 using C60 based biocompatible platform and enzyme functionalized Pt nanochains tracing tag. Biosensors and Bioelectronics. 2013;49:485–91. doi: 10.1016/j.bios.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Deng J, Fang LC, Yu KK, Huang H, Jiang LL, Liang WB, Zheng JS. A novel electrochemical DNA biosensor based on HRP-mimicking hemin/G-quadruplex wrapped GOx nanocomposites as tag for detection of Escherichia coli O157:H7. Biosensors and Bioelectronics. 2015;63:1–6. doi: 10.1016/j.bios.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 17.Tang DP, Hou L, Niessner R, Xu MD, Gao ZQ, Knopp D. Multiplexed electrochemical immunoassay of biomarkers using metal sulfide quantum dot nanolabels and trifunctionalized magnetic beads. Biosensors and Bioelectronics. 2013;46:37–43. doi: 10.1016/j.bios.2013.02.027. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X, Qi B, Li Y, Zhang S. Amplified electrochemical aptasensor for thrombin based on bio-barcode method. Biosensors and Bioelectronics. 2009;25:259–62. doi: 10.1016/j.bios.2009.06.026. [DOI] [PubMed] [Google Scholar]
- 19.Kong FY, Xu BY, Xu JJ, Chen HY. Simultaneous electrochemical immunoassay using CdS/DNA and PbS/DNA nanochains as labels. Biosensors and Bioelectronics. 2013;39:177–82. doi: 10.1016/j.bios.2012.07.023. [DOI] [PubMed] [Google Scholar]
- 20.Li X, Liu J, Zhang S. Electrochemical analysis of two analytes based on a dual-functional aptamer DNA sequence. Chemical Communications. 2010;46:595–7. doi: 10.1039/b916304b. [DOI] [PubMed] [Google Scholar]
- 21.Liang WB, Fan CC, Zhuo Y, Zheng YN, Xiong CY, Chai YQ, Yuan R. Multiparameter analysis-based electrochemiluminescent assay for simultaneous detection of multiple biomarker proteins on a single Interface. Analytical Chemistry. 2016;88:4940–8. doi: 10.1021/acs.analchem.6b00878. [DOI] [PubMed] [Google Scholar]
- 22.Chen AY, Gui GF, Zhuo Y, Chai YQ, Xiang Y, Yuan R. Signal-off electrochemiluminescence biosensor based on phi29 DNA polymerase mediated strand displacement amplification for microRNA detection. Analytical Chemistry. 2015;87:6328–34. doi: 10.1021/acs.analchem.5b01168. [DOI] [PubMed] [Google Scholar]
- 23.Zhang P, Wu XY, Yuan R, Chai YQ. “Off-On” electrochemiluminescent biosensor based on DNAzyme-assisted target recycling and rolling circle amplifications for ultrasensitive detection of microRNA. Analytical Chemistry. 2015;87:3202–7. doi: 10.1021/ac504455z. [DOI] [PubMed] [Google Scholar]
- 24.Zhang B, Liu BQ, Tang DP, Niessner R, Chen GN, Knopp D. DNA-based hybridization chain reaction for amplified bioelectronic signal and ultrasensitive detection of proteins. Analytical Chemistry. 2012;84:5392–9. doi: 10.1021/ac3009065. [DOI] [PubMed] [Google Scholar]
- 25.Guo JJ, Wang JC, Zhao JQ, Guo ZL, Zhang YZ. Ultrasensitive multiplexed immunoassay for tumor biomarkers based on DNA hybridization chain reaction amplifying signal. Applied Materials & Interfaces. 2016;8:6898–904. doi: 10.1021/acsami.6b00756. [DOI] [PubMed] [Google Scholar]
- 26.Wang D, Lu CH, Willner I. From cascaded catalytic nucleic acids to enzyme-DNA nanostructures: controlling reactivity, sensing, logic operations, and assembly of complex structures. Chemical Reviews. 2014;114:2881–941. doi: 10.1021/cr400354z. [DOI] [PubMed] [Google Scholar]
- 27.Wang D, Lu CH, Liu XQ, Freage L, Willner I. Amplified and multiplexed detection of DNA using the dendritic rolling circle amplified synthesis of DNAzyme reporter units. Analytical Chemistry. 2014;86:1614–21. doi: 10.1021/ac4033033. [DOI] [PubMed] [Google Scholar]
- 28.Zhao M, Zhuo Y, Chai YQ, Yuan R. Au nanoparticles decorated C60 nanoparticle-based label-free electrochemiluminesence aptasensor via a novel “on-off-on” switch system. Biomaterials. 2015;52:476–83. doi: 10.1016/j.biomaterials.2015.02.058. [DOI] [PubMed] [Google Scholar]
- 29.Hermanson GT. Bioconjugate Techniques. Waltham, USA: Elsevier Inc; 2013. [Google Scholar]
- 30.Hu K, Liu P, Ye S, Zhang S. Ultrasensitive electrochemical detection of DNA based on PbS nanoparticle tags and nanoporous gold electrode. Biosensors and Bioelectronics. 2009;24:3113–9. doi: 10.1016/j.bios.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 31.Hao Q, Shan XN, Lei JP, Zhang Y, Yang QH, Ju HX. A wavelength-resolved ratiometric photoelectrochemical technique: design and sensing applications. Chemical Science. 2016;7:774–80. doi: 10.1039/c5sc03336e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park S, Taton T, Mirkin C. Array-based electrical detection of DNA with nanoparticle probes. Science. 2002;295:1503–6. doi: 10.1126/science.1067003. [DOI] [PubMed] [Google Scholar]
- 33.Wang HJ, Bai LJ, Chai YQ, Yuan R. Synthesis of multi-fullerenes encapsulated palladium nanocage, and its application in electrochemiluminescence immunosensors for the detection of streptococcus suis serotype 2. Small. 2014;10(9):1857–65. doi: 10.1002/smll.201303594. [DOI] [PubMed] [Google Scholar]
- 34.Liu M, Wang ZY, Zong SF, Chen H, Zhu D, Wu L, Hu GH, Cui YP. SERS detection and removal of Mercury(II)/Silver(I) using oligonucleotide-functionalized core/shell magnetic silica sphere@Au nanoparticles. Applied Materials & Interfaces. 2014;6:7371–9. doi: 10.1021/am5006282. [DOI] [PubMed] [Google Scholar]
- 35.Ge JP, Wang J, Zhang HX, Wang X, Peng Q, Li YD. Orthogonal PbS nanowire arrays and networks and their Raman scattering behavior. Chemistry. 2005;11:1889–94. doi: 10.1002/chem.200400633. [DOI] [PubMed] [Google Scholar]
- 36.Malikov EY, Altay MC, Muradov MB, Akperov OH, Eyvazova GM, Puskas R, Madarasz D, Kukovecz A, Konya Z. Synthesis and characterization of CdS nanoparticle based multiwall carbon nanotube-maleic anhydride-1-octene nanocomposites. Physica E: Low-dimensional Systems and Nanostructures. 2015;69:212–8. [Google Scholar]
- 37.Kim GY, Son A. Development and characterization of a magnetic bead-quantum dot nanoparticles based assay capable of Escherichia coli O157:H7 quantification. Analytica Chimica Acta. 2010;677:90–6. doi: 10.1016/j.aca.2010.07.046. [DOI] [PubMed] [Google Scholar]
- 38.Gehring AG, Irwin PL, Reed SA, Tu SI, Andreotti PE, Akhavan-Tafti H, Handley RS. Enzyme-linked immunomagnetic chemiluminescent detection of Escherichia coli O157:H7. Journal of immunological methods. 2004;293:97–106. doi: 10.1016/j.jim.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 39.Gehring AG, Patterson DL, Tu SI. Use of a Light-addressable potentiometric sensor for the detection of Escherichia coli O157:H7. Analytical Biochemistry. 1998;258(2):293–8. doi: 10.1006/abio.1998.2597. [DOI] [PubMed] [Google Scholar]
- 40.Deng MY, Fratarnico PM. A Multiplex PCR for rapid identification of shiga-like toxin-producing Escherichia coli O157:H7 isolated from foods. Journal of food protection. 1996;59(6):570–6. doi: 10.4315/0362-028X-59.6.570. [DOI] [PubMed] [Google Scholar]
- 41.Sungkanak U, Sappat A, Wisitsoraat A, Promptmas C, Tuantranont A. Ultrasensitive detection of Vibrio cholerae O1 using microcantilever-based biosensor with dynamic force microscopy. Biosensors and Bioelectronics. 2010;26:784–9. doi: 10.1016/j.bios.2010.06.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.