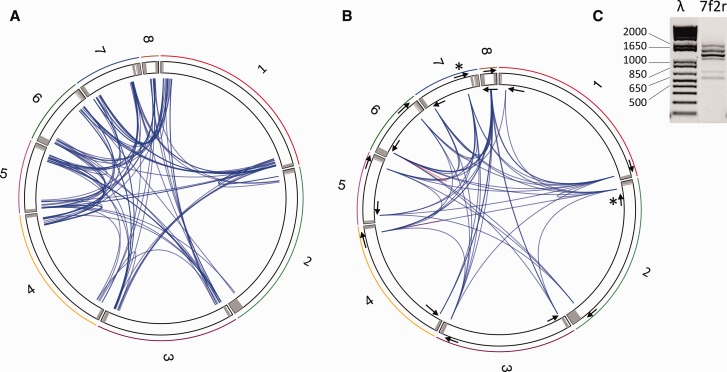
Fig. 5.—
Connexion among contigs identified by (A) mapping paired-end reads on the LbFV contigs masked for repeated sequence. The eight LbFV contigs are arbitrarily ranked according to their length and represented within a circular chromosome. Grey areas indicate genomic regions homologous to the hrs motif. The blue lines represent the mapping of paired-end reads on different contigs extremities. In total 96 blue lines are represented corresponding to 96 paired Illumina reads. (B) PCR amplification. The 16 PCR primers used in this experiment are symbolically represented by the arrows (not scale). 51 out of 120 primer combinations gave a positive PCR and are represented by a blue line connecting contig extremities. The red lines indicate two cases where the PCR was positive with a single primer. (C) The PCR amplifications often gave several bands. As an example, the PCR product obtained with the 7f and 2r primers (identified in fig. 5B by stars) is shown. The full set of gel images is provided in supplementary figure S2, Supplementary Material online. λ, molecular weight marker; 7f2r, PCR product.