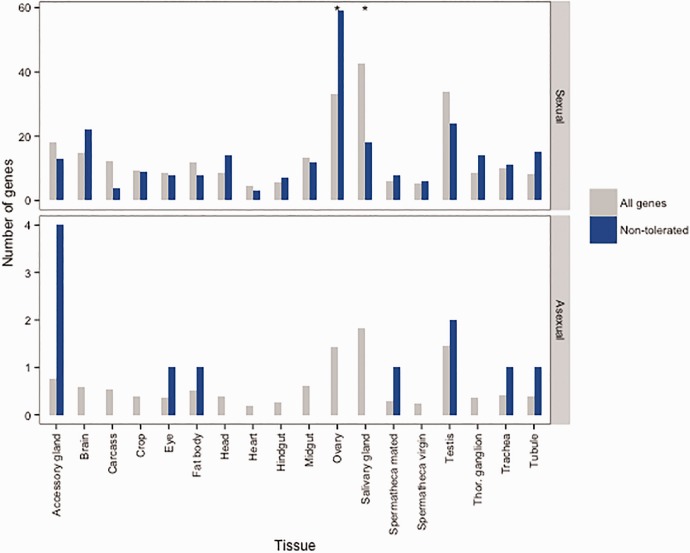
Fig. 3.—
Deleterious variants in the Leptopilina clavipes genome are overrepresented in reproductive tissues. Deleterious (non-tolerated) variants were identified using SIFT and the orthologs of the genes in which they were found were searched for in the genome of Drosophila melanogaster. The tissue in which each of these orthologs show highest expression was identified in Flyatlas (Chintapalli et al. 2007) and is shown in blue for asexual and sexual L. clavipes lineages. The distribution of tissues with most abundant expression for all genes in Flyatlas is shown in grey. Significant Fisher exact P values following FDR correction are indicated with an asterisk.