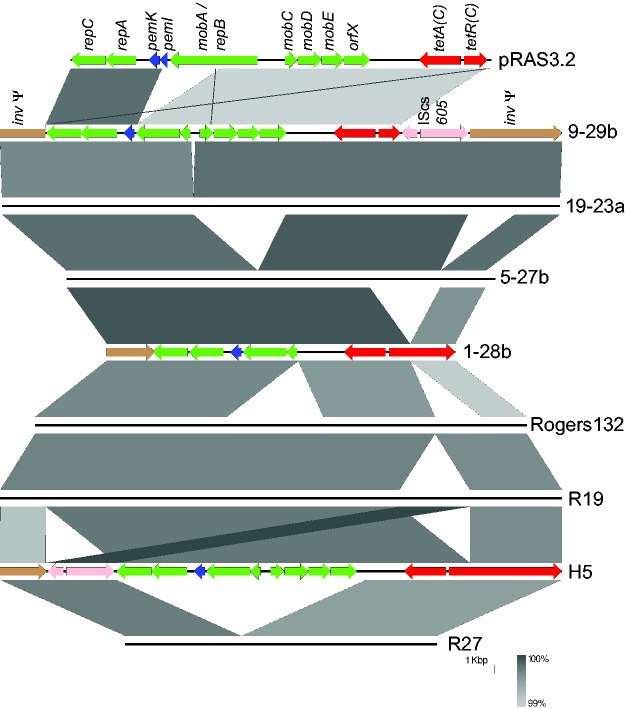
Fig. 4.—
Variations of organization of TetC-carrying islands, compared against the putative ancestor pRAS3.2. Islands/plasmids are shown as lines, with the grey bars indicating nucleotide identity between island pairs. The top line represents pRAS3.2, followed by the island from isolate 9-29b (both of these are annotated with their encoded genes), which reflects the gene organization seen in strains 8-29b, 11-17a, 11-17b, 5-22b, 30-22b, 15-27b, 17-23a, 3-29b, 4-29b, and 22-22b (the latter from partial sequence information only) as well as the US strains R19 and R24. The island is inserted within an invasin gene (inv), splitting it into two pieces and rendering it a pseudogene (brown, left and right of the figure). The CDSs encoding plasmid-related Rep and Mob proteins (green), PemK (blue), and an insertion sequence (IScs605 with two transposase tnp genes; pink) are found adjacent to the tet operon (red). Strain 19-23a has a small deletion relative to this within the mob operon. Strain 5-27b has two deletions: comprising much of the mob operon and part of inv. Strain 1-28b (annotated) has these deletions and a further one which creates a fusion protein between the tetR (C) regulator and the C terminal portion of inv, indicated by the extended red CDS on the right. The organization of the US strain Tet-islands is shown in the lower part of the figure, with islands from Rogers132, R19 (full length island, also representing R24), H5 (annotated, showing translocation of IScs605 and tetR(C)-inv fusion), and R27 (reduced island, representing the gene organization in R1, R16, R22, R28, and H7) (Joseph et al. 2016). All nucleotide identities are in excess of 99% (see scale). Image produced using Easyfig (Sullivan et al. 2011).