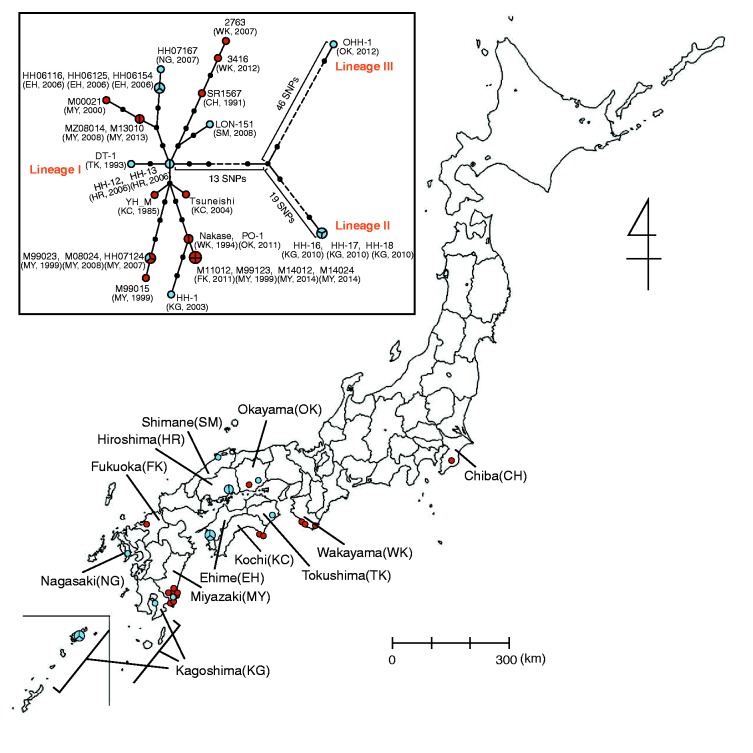
Fig. 1.—
Geographic distribution of Rickettsia japonica analyzed in this study and associated phylogenetic relationships. Isolation locations for the 31 analyzed strains are shown (see Supplementary fig. S2, Supplementary Material online, for more precise information of the strains isolated in Miyazaki prefecture). Clinical and tick isolates are indicated by red and blue circles, respectively. Larger circles that are divided into two or three parts indicate two or three strains isolated from ticks collected in the same location (within a few kilometers) on the same day. The names of the prefecture where the strains were isolated are indicated. The abbreviations of each prefecture are shown in parentheses. In the box, a phylogenetic tree of the 31 R. japonica strains is shown. The tree was constructed by using 112 SNP sites that were identified among the 31 strains. Clinical and tick isolates are indicated by red and blue circles, respectively. Larger circles that are divided into two, three, or four parts represent 2-, 3-, or 4-member strain sets with identical genome sequences in terms of SNPs, respectively. Small dots represent hypothetical intermediates, and the dot-to-dot distance corresponds to one SNP difference. The years and locations of strain isolation are shown in parentheses. Among the three lineages identified, Lineage II contains strains HH-16/17/18, and Lineage III contains one strain (OHH-1). Lineage I contains the other 27 strains. Among the 112 SNPs, we found no homoplasic SNP (the same mutation that occurred in different lineages independently or in parallel).