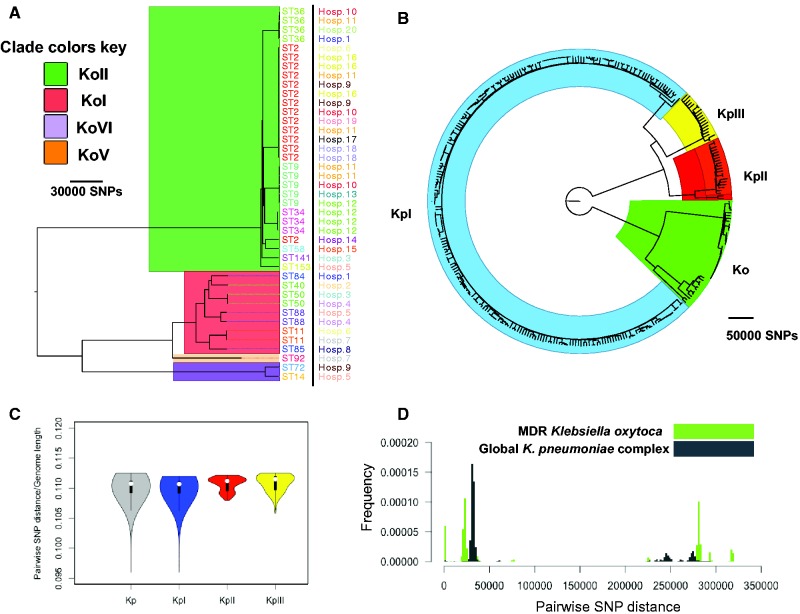
Fig. 1.—
(A) Maximum Likelihood phylogenetic tree for the MDR Klebsiella oxytoca collection. Hosp. and ST stand for hospital of isolation and sequence type, respectively. The background colours signify the major phylogroups of K. oxytoca. (B) Combined phylogenetic tree of the global Klebsiella pneumoniae complex (Kp) collection and the MDR K. oxytoca (Ko) collection. The background colours correspond to the species within the global K. pneumoniae complex collection. KpI, KpII and KpIII correspond to K. pneumoniae, Klebsiella quasipneumoniae, and Klebsiella variicola, respectively. (C) Distribution of SNP distances for pairs of K. oxytoca and the whole K. pneumoniae complex (Kp) and each species within the K. pneumoniae complex. The values have been divided by the genome length of reference genome used for mapping. The boxes give the interquartile range and the whiskers indicate the boundary of 1.5 times the interquartile range. The white marker shows the median of the data and the coloured area is the probability density of the data at different values. (D) Distributions of pairwise SNP distances for the K. oxytoca collection and the global K. pneumoniae complex.