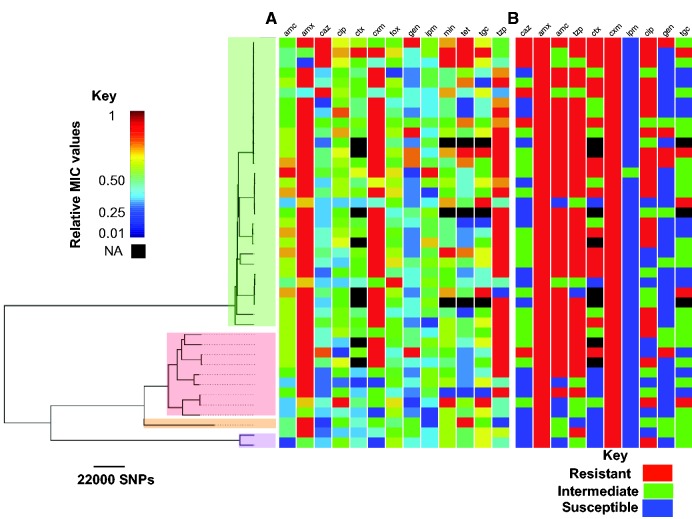
Fig. 4.—
(A) Distribution of MICs and (B) resistance phenotype across the phylogenetic tree. The clade colours are the same as in figure 1A and show K. oxytoca phylogroups. Abbreviations of the antibiotics are: amoxicillin (amx), cefuroxime (cxm), amoxicillin–clavulanate (amc), cefotaxime (ctx), cefoxitin (fox), imipenem (ipm), piperacillin–tazobactam (tzp), ciprofloxacin (cip), ceftazidime (caz), gentamicin (gen), tigecycline (tgc), minocycline (min) and tetracycline (tet). We normalized the MICs for each antibiotic such that the maximum and minimum MICs take values 1 and 0, respectively. Note that we show the phenotypes only for antibiotics for which clinical breakpoints were defined.