Abstract
Many species are not completely reproductively isolated, resulting in hybridization and genetic introgression. Organellar genomes, such as those derived from mitochondria (mtDNA) and chloroplasts, introgress frequently in natural systems; however, the forces shaping patterns of introgression are not always clear. Here, we investigate extensive mtDNA introgression in western chipmunks, focusing on species in the Tamias quadrivittatus group from the central and southern Rocky Mountains. Specifically, we investigate the role of selection in driving patterns of introgression. We sequenced 51 mtDNA genomes from six species and combine these sequences with other published genomic data to yield annotated mitochondrial reference genomes for nine species of chipmunks. Genomic characterization was performed using a series of molecular evolutionary and phylogenetic analyses to test protein-coding genes for positive selection. We fit a series of maximum likelihood models using a model-averaging approach, assessed deviations from neutral expectations, and performed additional tests to search for codons under the influence of selection. We found no evidence for positive selection among these genomes, suggesting that selection has not been the driving force of introgression in these species. Thus, extensive mtDNA introgression among several species of chipmunks likely reflects genetic drift of introgressed alleles in historically fluctuating populations.
Keywords: Tamias, mitochondrial genomics, phylogenomics, molecular evolution, selection, introgression
Introduction
Interspecific hybridization occurs frequently in natural systems. Approximately 23% of animals exhibit mitochondrial DNA (mtDNA) polyphyly (Funk and Omland 2003), and approximately 10% of species are estimated to hybridize (Gray 1972; Mallet 2005). Although hybridization may promote genetic homogenization by collapsing incipient lineages into hybrid swarms (Taylor et al. 2006; Behm et al. 2010; Gilman and Behm 2011), it is becoming clear that gene flow may not prevent lineage divergence (Wu 2001; Pinho and Hey 2010). Models detailing this process are captured by the overarching term “divergence-with-gene-flow” (Rice and Hostert 1993) and describe the speciation process as a more dynamic accumulation of reproductive barriers over time.
One of the most strongly recurring patterns in hybridizing systems is that organellar genomes (i.e., the mitochondrion and chloroplast) often introgress. Indeed, much of the evidence for hybridization in animals derives from studies of mtDNA introgression as revealed through topological discordance, which has been documented in many groups, including lizards, amphibians, birds, and mammals (Funk and Omland 2003; Mallet 2005; McGuire et al. 2007; Bossu and Near 2009; Ryan et al. 2009; Johanet et al. 2011; Rheindt and Edwards 2011). The high frequency of mitochondrial introgression may result from selection on haplotypes in a novel genetic or ecological background (Hedrick 2013; Llopart et al. 2014), and adaptive introgression of mtDNA has been inferred in instances of frequent mtDNA introgression (Melo-Ferreira et al. 2014).
Alternatively, genetic drift can promote haplotype fixation in small populations either through chance or in concert with selection following introgression (Ballard and Whitlock 2004). A special case of genetic drift is allele surfing, in which rare alleles present on the front of expanding populations may become fixed (Edmonds et al. 2004; Hallatschek et al. 2007; Excoffier and Ray 2008; Hallatschek and Nelson 2008); such “surfing” alleles can either be deleterious, neutral, or advantageous (Klopfstein et al. 2006; Travis et al. 2007; Excoffier et al. 2009). A potential outcome of allelic surfing is the introgression of resident alleles into the expanding population (Currat and Excoffier 2004; Currat et al. 2008). This is expected to result in an asymmetric pattern of introgression, and empirical results provide support for this prediction; 36 of the 44 studies examined by Currat et al. (2008) involve allelic introgression from the resident population into the invading population. In this case, the extent of introgression appears to be governed jointly by allelic fitness, the rate of migration, and stochastic demographic effects (Petit and Excoffier 2009).
Here, we focus on mitochondrial introgression in chipmunks (Sciuridae: Tamias), a diverse group of ground squirrels composed of 25 species (Thorington and Hoffman 2005). Of these, one species is restricted to eastern Asia (T. sibiricus; subgenus Eutamias), and one is restricted to eastern North America (T. striatus; subgenus Tamias). The remaining 23 species in the subgenus Neotamias (but see Piaggio and Spicer 2000, 2001; Patterson and Norris 2016) are distributed throughout western North America. Assignment to species has relied on variation in the male genital bone, the baculum or os penis, with variation of other phenotypic characters (e.g., pelage and body size) showing considerable overlap among species. The entire subgenus in western North America has been subject to periodic range shifts during the Pleistocene as divergence was occurring. Therefore, many species pairs are likely to have experienced several periods of ephemeral contact over the past ∼2.5 Myr, punctuated by periods of isolation (Sullivan et al. 2014).
Previous work in chipmunks (reviewed in Sullivan et al. 2014) has documented extensive mtDNA introgression. Of 1871 individuals that have been sequenced (from nearly all species), nearly 300 exhibit mtDNA that has been inferred to be introgressed, and these introgressions involve 40% of the species in the genus. This has been most thoroughly documented in three cases. First, several studies have documented asymmetric introgression of mtDNA between two subspecies of red-tailed chipmunk (T. ruficaudus ruficaudus into T. r. simulans; Good and Sullivan 2001; Hird and Sullivan 2009; Hird et al. 2010), and a small number of putative F1 hybrids have been inferred from variation at microsatellite loci (Hird and Sullivan 2009). Second, several other studies have documented asymmetric introgression of T. ruficaudus mtDNA genome into the yellow-pine chipmunk (T. amoenus) (Good et al. 2003, 2008, 2015; Reid et al. 2010, 2012), even though these species are rather distantly related (Reid et al. 2012). Third, more recent work has demonstrated widespread introgression of mtDNA between several pairs of species in the T. quadrivitattus group, which has likely diverged within the past ∼1.7 Myr (Sullivan et al. 2014). In each of these cases, coalescent simulations have strongly supported the inference of mtDNA introgression (Reid et al. 2012).
In all cases, introgression appears to be asymmetric and may be mediated by differences in genital morphology, particularly of the baculum. The mating system of the genus and ecologies of introgressing species of Tamias are relevant and interesting. For the most part, the species are monestrous, and mating typically occurs when the female is in estrus on only 1 day per year (Callahan 1981). High rates of multiple paternity have been reported (for T. amoenus: Schulte-Hostedde et al. 2004), and there is evidence that sexual selection is important and may involve sperm competition (Schulte-Hostedde and Millar 2004). Further, the genus has long been known for providing classic examples of niche partitioning among broadly sympatric taxa that frequently results in altitudinal zonation (Heller 1971), generating narrow zones of paraparty (sometimes called contiguous allopatry). Indeed, many of the introgressions we have identified occur between ecologically differentiated taxa that only come into contact at habitat transitions (e.g., T. ruficaudus and T. amoenus at the transition between mesic and xeric forests in Montana and Idaho; Gambs 1965; Best 1993; T. umbrinus and T. dorsalis at the transition between sparse and dense Pinyon-Juniper forests in Nevada; Brown 1971).
The extensive, recurrent, and highly asymmetric nature of mtDNA introgression in Tamias is striking and occurs between ecologically differentiated taxa across a range of divergence times. The repeated nature of asymmetric introgression raises the question of whether these patterns are primarily driven by neutral processes or positive selection. Previous studies in chipmunks have relied upon a single mtDNA gene (cyt b) to detect introgression between species and, therefore, could not assess the molecular evolution of chipmunk mtDNA genome in the context of introgression. Here, we combine data from T. amoenus, T. ruficaudus, and T. striatus published by Bi et al. (2012) with complete mtDNA genomes from 51 individuals across the T. quadrivittatus group to characterize the evolutionary history of these differentially introgressed genomes. We use these data to assess the role of selection in explaining widespread mtDNA introgression in Tamias by explicitly testing for positive selection across the mtDNA genome. If positive selection is detected, we have evidence for an adaptive explanation for mtDNA introgression; if not, we conclude that demography likely explains the patterns of introgression in this system.
Materials and Methods
Biological Sampling
To date, approximately 1800 mtDNA cyt b sequences have been generated in chipmunks (reviewed in Sullivan et al. 2014), nearly 300 of which have introgressed haplotypes. We selected 56 chipmunks from taxa that have been shown to exhibit extensive mtDNA introgression (Good et al. 2003; Sullivan et al. 2014). These included T. ruficaudus ruficaudus, T. r. simulans, T. amoenus luteiventris, T. a. canicaudus, and T. striatus (one each, which were sequenced as part of a separate study; Bi et al. 2012), as well as 51 individuals from the T. quadrivittatus group (T. canipes, T. rufus, T. quadrivittatus, T. cinereicollis, T. dorsalis, and T. umbrinus; see supplementary table S2, Supplementary Material online for collection localities). Samples were selected from throughout the species range when possible. The T. quadrivittatus-group sample included both introgressed and nonintrogressed individuals. DNA was isolated from heart or liver tissue using Qiagen DNEasy extraction kits and eluted in ∼50 μL of 10 mM Tris-Cl. Extractions were stored at −20 °C prior to use. All vouchers for the T. quadrivittatus group are deposited at the Denver Museum of Nature & Science.
Obtaining a Draft Tamias Mitochondrial Genome
Prior to this study, no reference mitochondrial genome was available for Tamias. We therefore generated a preliminary reference via primer walking and Sanger sequencing. In order to design primers (using Primer3; Untergasser et al. 2007), we generated a consensus sequence from the Eurasian red squirrel, (Sciurus vulgaris, Reyes et al. 2000) and the edible dormouse (Glis glis; Reyes et al. 1998). Pairwise combinations of primers for PCR were utilized in 50 μL amplifications consisting of 39.3 μL of ddH2O, 5 μL of 10× buffer, 1 μL of 10 mM dNTPs, 1.5 μL of 50 mM MgCl2, 1 μL of a 100 μM solution of each primer, 0.2 μL of polymerase, and 2 μL of genomic DNA. Amplification was performed on a BioRad MyCycler with the following parameters: 94 °C for 2:00 followed by 45 cycles of 94 °C for 0:30, 55 °C for 0:45, and 72 °C for a variable period depending on the size of the region amplified (assuming 1:00/1000bp). Each reaction was subject to a 5:00 final extension at 72 °C.
Amplicons were cleaned using Qiagen PCR purification kits. Purified amplicons were prepared for cycle sequencing in 8 μL reactions consisting of 2 μL Big Dye, 1.6 μL 100 1 μM primer solution, and 4.4 μL purified amplicon. Cycle sequencing was performed on the same thermocycler under the following conditions: 96 °C for 1:00, followed by 25 cycles of 96 °C for 0:15, 50 °C for 0:15, and 60 °C for 4:00. Sequencing reactions were cleaned using Sephadex spin columns, dried, and suspended in 10 μL of formamide for sequencing on an ABI 3130 capillary sequencer. A final set of at least 89 primer pairs, including some redundancy and approximately 200 bp of overlap between loci, was used to amplify and sequence a draft mitochondrial genome. The draft mitochondrial genome was checked for sequence quality and assembled into a working draft genome using the Sciurus mitochondrial genome as a backbone.
Targeted Capture and Mitochondrial Genome Assembly
The draft mitochondrial genome was included as part of a set of approximately 12,000 exons that were used as baits for a targeted capture experiment (see Bi et al. 2012 for details). Fifty-one samples across six species of chipmunks (Tamias canipes, T. rufus, T. quadrivittatus, T. cinereicollis, T. dorsalis, and T. umbrinus: the T. quadrivittatus group, fig. 1) were captured on an Agilent SureSelect 1M microarray using a previously described protocol (Bi et al. 2012). Samples were sequenced on two lanes of an Illumina HiSeq 2000 at the Vincent J. Coates Genome Sequencing Laboratory at the University of California, Berkeley. In addition, raw data from five additional individuals (T. r. ruficaudus, T. r. simulans, T. a. luteiventris, T. a. canicaudus, and T. striatus) captured on the same microarray platform as part of another study (Bi et al. 2012) were included in this analysis. Libraries were cleaned using SeqyClean (https://github.com/ibest/seqyclean) in order to remove low-quality reads (<50 bp post-trimming), low-quality bases (minimum base quality of 20), and residual Illumina adapter sequences. Additionally, reads were de-duplicated, and any reads that overlapped were merged using FLASH (Magoč and Salzberg 2011).
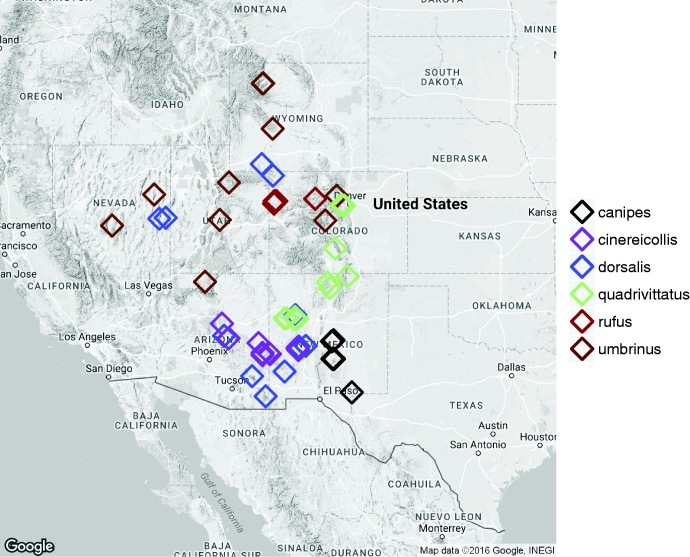
Fig. 1.—
Sampling localities of the 51 T. quadrivittatus group individuals sequenced as part of this study. The map was generated with ggmap (Kahle and Wickham 2013) using the Google Maps API. Points are slightly shifted for clarity in showing overlapping sampling localities.
Cleaned reads were used as the starting point for de novo assembly using Assembly by Reduced Complexity (ARC; Hunter et al. 2015; current version available from http://ibest.github.io/ARC). Briefly, ARC identifies reads that are similar to a target of interest, places these reads into a reduced-representation pool, and performs a de novo assembly on the reduced pool of reads. This process is then iterated until no new reads are incorporated. This approach reduces assembly time by limiting the number of comparisons that must be made during assembly, as compared with de novo assembly of the entire library, and reduces biases that may be introduced through calling variants relative to a divergent reference in a different species (Hunter et al. 2015). The S. vulgaris mitochondrial genome was used as the target sequence for assembly of a randomly selected T. canipes mitochondrial genome. This mitochondrial genome was then used as the target for all other assemblies in order to reduce reference bias and expedite assembly.
Following assembly, the resulting contig(s) were oriented to a common start site (tRNA-Phenylalanine/12S rRNA) by mapping and manual reorientation using Geneious Pro v6.1.7 (Kearse et al. 2012). Protein-coding genes and rRNAs were identified by free-end alignment to the Sciurus mtGenome and through annotation relative to Sciurus and Mus in Geneious Pro. A representative mitochondrial genome from each of the nine species (including the two subspecies of T. ruficaudus and two of T. amoenus) was used to calculate nucleotide frequencies using Geneious Pro v. 6.1.7. Final mtGenome sequences are deposited in Genbank under accession numbers KY070142-KY070197. A visualization of the annotated version of the draft genome is included as part of the supplementary material (supplementary fig. S1, Supplementary Material online).
In order to provide an independent validation for the de novo assemblies of the mitochondrial genomes, the lysine transfer RNA (tRNA-Lys) was extracted from each genome. This tRNA has an interesting evolutionary history within Mammalia (Dorner et al. 2001; see supplementary fig. S2 and accompanying description). A representative from each species and subspecies was annotated, extracted, and transcribed using Geneious Pro v6.1.7. The secondary structure was generated using the RNAfold web server (Hofacker 2003). The secondary structure of the Mus musculus tRNA-Lys was also visualized using the same approach.
Phylogenetic Analyses
The 13 protein coding and two rRNA loci from both datasets were aligned using Mafft v6.86b and the G-INS-i (global homology) algorithm (Katoh and Toh 2008). A model of nucleotide sequence evolution was inferred for each locus using the decision theoretic approach implemented in DT-ModSel (Minin et al. 2003). We chose to use this approach for model selection because decision-theoretic approaches have been shown to select simpler models that minimize branch-length error (and therefore topological error) relative to other approaches (Minin et al. 2003; Abdo et al. 2005; Luo et al. 2010). Maximum-likelihood phylogenetic estimation under the inferred model were performed using Garli v1.0, with a score threshold of 0.01 and a requirement of 500,000 stable generations prior to termination (Zwickl 2006). We performed 10 replicate searches, and convergence among searches was assessed by identification of identical or highly similar trees and similar log-likelihood scores. Trees from a preliminary dataset were used to assess topological incongruence among genes using the conservative Shimodaira–Hasegawa (Shimodaira and Hasegawa 1999) test implemented in PAUP* v4.10b (Swofford 2003). These tests used the model of sequence evolution selected by DT-ModSel and calculated likelihoods using RELL bootstrapping with 1000 replicates. The tree generated from each locus was compared with the tree generated using the mitochondrial genome, which was included in the set of alternative topologies estimated from each individual locus.
A maximum clade-credibility tree for the entire mitochondrial genome of the complete dataset was generated from a posterior distribution of trees estimated using BEAST v1.8.0 (Drummond and Rambaut 2007; Drummond et al. 2012). Two identical runs were performed using a GTR + I+Γ model of nucleotide sequence evolution, a birth-death tree prior, and an uncorrelated lognormal relaxed molecular clock. Chains were run for 50 million generations with samples taken every 5000. Convergence between runs was assessed through visual inspection of traces, comparison of mean parameter estimates, and verification that the effective sampling size (ESS) of parameters were at least 200 or greater. The posterior was summarized into a maximum clade credibility tree after removing 10% of samples as a burn-in using TreeAnnotator v1.8.0, with median node heights and a nodal probability cutoff of 0.5.
Tests for Selection
Because mitochondrial introgression is rampant in T. dorsalis (Sullivan et al. 2014), we utilized two datasets. To reduce the influence of interspecific polymorphisms, the first dataset consisted of one T. canipes, T. cinereicollis, T. quadrivittatus, T. rufus, T. umbrinus, T. amoenus, and T. ruficaudus plus T. striatus. The second consisted of five T. dorsalis with an introgressed mitochondrial haplotype (as identified using the mtGenome tree) plus a T. canipes and T. rufus, both of which are monophyletic with respect to their mitochondrial genome, and was used to test whether ω (dN/dS) differs among introgressed T. dorsalis. Additionally, we removed ND6 from analysis due to its disparate base composition, and we removed any codons that were part of multiple genes.
We inferred patterns of selection using PAML v4.6 (Yang 2007) and a range of site models (M0, M1a, M2a, M3, M4, M5, M6, M7, M8, and M8a). The simplest of these models is M0, which assumes a single ω across all sites. The other models estimate the proportion of sites assigned to a range of site classes when the ω corresponding to these classes is constrained in some fashion. For example, M1a estimates the proportion of sites assigned to two classes, one where ω is fixed at one and another where ω < 1. Alternatively, model M2a estimates the proportion of sites assigned to three classes, one where ω = 1, another where ω < 1, and a third where ω > 1. These models, and the others, are described in greater detail within the PAML manual.
Two approaches are commonly used to test for selection using these sets of models. The first is to select among the models using an information theoretic criterion, typically the AIC. The second is to perform a series of likelihood-ratio tests (LRT) among nested models that allow sites to be placed into a class with a ω greater than one. Three likelihood-ratio tests were performed: M1a vs. M2a, M7 vs. M8, and M8 vs. M8a. Models M2a and M8 allow for assignment of codons to a site class where ω is greater than one and, therefore, allow for the detection of positive selection. M8a includes a site class where ω = 1 instead of being allowed to vary. The null distribution of the test statistic using these two models is a mixture and therefore differs from the standard chi-square distribution used to calculate p-values for the other two comparisons (see details in the PAML manual).
However, the LRT approach is not applicable for non-nested models (e.g., M3, M4, M5, and M6). In order to accommodate uncertainty in model choice into our assessment of selection, we implemented a model-averaging approach (Sullivan and Joyce 2005). Although some authors (Posada and Buckley 2004), have advocated model averaging based on δAIC, the posterior probabilities calculated by Bayesian approaches represent a more explicit treatment of the model as a random variable (Sullivan and Joyce 2005). The posterior probability of a model can be approximated as:
where the summation in the denominator is across all m models in the candidate pool and
Here, ki is the number of free parameters in model i, and n is the sample size (Raftery 1995; this is usually approximated by sequence length). This approach assumes uniform (or vague; Schwarz 1978) prior probabilities across models. Furthermore, because lnL is typically calculated at its highest point (and therefore estimates a joint rather than marginal probability), this use of the BIC to approximate posterior probabilities assumes that the joint MLEs approximate the marginal likelihoods. Evans and Sullivan (2011) used reversible-jump MCMC to estimate model probabilities directly and assessed the usefulness of the BIC as an approximation of probabilities for models of nucleotide substitution. They found that the approximation works well when there is much information in the data regarding model preference (Evans and Sullivan 2011). Therefore, we implemented the above approach in a Python script (available at https://github.com/bricesarver/codemlMA) to perform LRTs, calculate BICs, and approximate posterior probabilities of the models available in PAML. We then used this approach to derive model-averaged estimates of parameters (κ and ω). Additionally, for the T. dorsalis dataset, we evaluated additional models to test for lineage-specific positive selection: a null model assuming one ω across all branches and a model assuming a different ω for T. dorsalis lineages. These models were compared using an LRT to assess significance.
Subsequently, we performed several additional tests to look for positive selection across protein-coding loci in the T. quadrivittatus group. Five models were fit to attempt to identify codons under selection: SLAC (Single-Likelihood Ancestor Counting; Kosakovsky Pond and Frost 2005a,b), FEL (Fixed Effects Likelihood; Kosakovsky Pond and Frost 2005a,b), IFEL (Internal Fixed Effects Likelihood; Kosakovsky Pond et al. 2006), REL (Random Effects Likelihood; Kosakovsky Pond and Frost 2005a,b), and FUBAR (Fast, Unconstrained Bayesian AppRoximation; Murrell et al. 2013). All analyses were performed using the Datamonkey web server (Kosakovsky Pond and Frost 2005a), a publicly accessible front end to a cluster computing system running HyPhy (Kosakovsky Pond et al. 2005). Furthermore, we performed pairwise McDonald–Kreitman tests (McDonald and Kreitman 1991), using DnaSP v5 (Librado and Rozas 2009) using the same regions as above for five species (T. quadrivittatus, T. cinereicollis, T. rufus, T. canipes, and T. umbrinus) plus T. amoenus. Finally, we calculated Tajima’s D (Tajima 1989), a population genetic statistic commonly used to test for neutrality based on the site frequency spectrum, for all sites, synonymous sites, and nonsynonymous sites.
Results
Characteristics of the Chipmunk Mitochondrial Genome
Chipmunk mtDNA genomes exhibit syntenic conservation with other mammalian mtDNA genomes. 36% of the positive strand is composed of cytosine or guanine nucleotides. This strand also has a 46:54 purine:pyrimidine bias across all species. Nucleotide frequencies are similar across species (table 1). Lengths of protein-coding genes are largely conserved, but there is some interspecific length variation in the control region and ribosomal RNAs associated with indels. Several genes are missing one or two nucleotides that make up the final stop codon. In Sciurus and other mammals, the stop codon appears to be completed through the polyadenylation of pre-mRNAs (Reyes et al. 2000; Chang and Tong 2012). Tamias also appears to require this modification, as several exons do not end in an appropriate mammalian stop codon. Each of the T. quadrivittatus group species show approximately 6% uncorrected sequence divergence from one another and approximately 15% divergence from T. striatus, the outgroup (table 2). Distributions of variable sites across codon positions for each gene (table 3) are typical, with many third-position sites most variable, followed by first-positions, with second-position sites being the most conserved.
Table 1.
Descriptive Characteristics of Chipmunk Mitochondrial Genomes
| Species | ID | A | C | G | T | Total Length | G/C | A/G | C/T |
|---|---|---|---|---|---|---|---|---|---|
| T. canipes | DZTM.156 | 5556 | 4001 | 1995 | 5000 | 16,552 | 0.36 | 0.46 | 0.54 |
| T. cinereicollis | DZTM.230 | 5531 | 4008 | 2021 | 4989 | 16,549 | 0.36 | 0.46 | 0.54 |
| T. dorsalis | DZTM.713 | 5501 | 3991 | 2023 | 4986 | 16,501 | 0.36 | 0.46 | 0.54 |
| T. quadrivittatus | DZTM.700 | 5548 | 3997 | 2010 | 4994 | 16,549 | 0.36 | 0.46 | 0.54 |
| T. rufus | DZTM.572 | 5530 | 4002 | 2012 | 5000 | 16,544 | 0.36 | 0.46 | 0.54 |
| T. umbrinus | DZTM.256 | 5538 | 4009 | 2016 | 4986 | 16,549 | 0.36 | 0.46 | 0.54 |
| T. a. canicaudus | JMG102 | 5534 | 3947 | 2010 | 5061 | 16,552 | 0.36 | 0.46 | 0.54 |
| T. a. luteiventris | JMS297 | 5535 | 3965 | 2017 | 5028 | 16,545 | 0.36 | 0.46 | 0.54 |
| T. r. ruficaudus | JMG16 | 5517 | 3991 | 2024 | 5024 | 16,556 | 0.36 | 0.46 | 0.54 |
| T. r. simulans | JMS123 | 5512 | 3975 | 2029 | 5037 | 16,553 | 0.36 | 0.46 | 0.54 |
| T. striatus | Stri#2 | 5527 | 3793 | 2048 | 5185 | 16,553 | 0.35 | 0.46 | 0.54 |
| Mean | 5530 | 3971 | 2019 | 5026 | 16,546 | 0.36 | 0.46 | 0.54 | |
| Standard Deviation | 16 | 62 | 13 | 58 | 15 | 0.00 | 0.00 | 0.00 |
Note.—A representative individual of each species is used and is identified by the ID column. Nucleotide frequencies, G/C content, and purine/pyrimidine content are listed relative to the positive strand.
Table 2.
Pairwise p-Distances Among Representative Mitochondrial Genomes for Each Species
| T. canipes | T. cinereicollis | T. dorsalis | T. quadrivittatus | T. rufus | T. umbrinus | T. amoenus canicaudus | T. amoenus luteiventris | T. ruficaudus ruficaudus | T. ruficaudus simulans | T. striatus | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| T. canipes | – | ||||||||||
| T. cinereicollis | 0.046 | – | |||||||||
| T. dorsalis | 0.048 | 0.017 | – | ||||||||
| T. quadrivittatus | 0.047 | 0.026 | 0.025 | – | |||||||
| T. rufus | 0.045 | 0.027 | 0.028 | 0.028 | – | ||||||
| T. umbrinus | 0.047 | 0.012 | 0.016 | 0.023 | 0.027 | – | |||||
| T. amoenus canicaudus | 0.061 | 0.059 | 0.062 | 0.061 | 0.060 | 0.060 | – | ||||
| T. amoenus luteiventris | 0.067 | 0.065 | 0.067 | 0.067 | 0.065 | 0.066 | 0.065 | – | |||
| T. ruficaudus ruficaudus | 0.06 | 0.059 | 0.061 | 0.061 | 0.058 | 0.059 | 0.034 | 0.065 | – | ||
| T. ruficaudus simulans | 0.062 | 0.059 | 0.061 | 0.060 | 0.058 | 0.059 | 0.022 | 0.065 | 0.031 | – | |
| T. striatus | 0.146 | 0.146 | 0.145 | 0.147 | 0.146 | 0.146 | 0.145 | 0.147 | 0.148 | 0.145 | – |
Table 3.
Distribution of Variable Sites Across Codon Positions for Each the Protein-Coding Genes
| Gene | 1st | 2nd | 3rd |
|---|---|---|---|
| ATP6 | 25 (19) | 1 (1) | 125 (93) |
| ATP8 | 8 (5) | 6 (4) | 28 (23) |
| COI | 38 (26) | 0 (0) | 302 (230) |
| COII | 23 (13) | 1 (1) | 120 (94) |
| COIII | 37 (25) | 16 (15) | 160 (120) |
| CYTB | 45 (32) | 9 (8) | 260 (215) |
| ND1 | 77 (73) | 29 (24) | 203 (162) |
| ND2 | 94 (73) | 58 (48) | 214 (175) |
| ND3 | 20 (17) | 5 (3) | 67 (55) |
| ND4 | 79 (54) | 26 (16) | 277 (216) |
| ND4L | 22 (18) | 3 (1) | 51 (40) |
| ND5 | 119 (87) | 49 (40) | 367 (278) |
| ND6 | 111 (107) | 105 (103) | 145 (136) |
| Sum | 698 (535) | 308 (264) | 2319 (1837) |
Note.—The numbers in parentheses are for comparisons excluding the outgroup (T. striatus).
Phylogenetic Inference
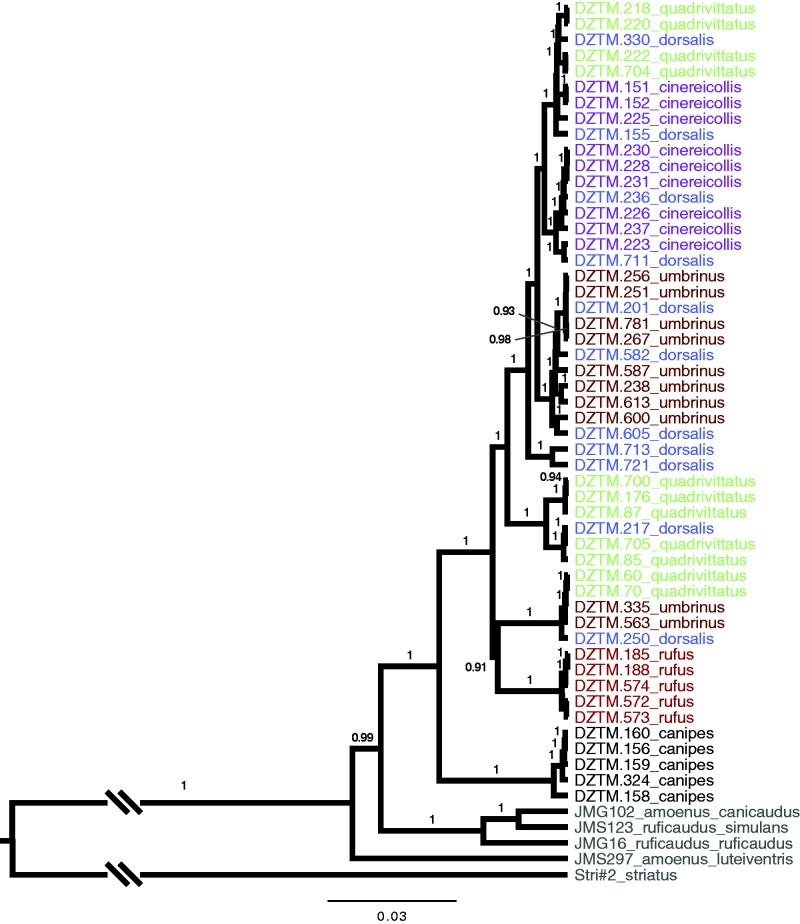
DT-ModSel selected the most parameter-rich model, GTR + I + Γ, for the complete mtGenome alignment. Phylogenetic analysis of complete mtDNA genomes is broadly congruent with the cytochrome b tree estimated in Sullivan et al. (2014) (fig. 2). The yellow-pine (T. amoenus) and red-tailed (T. ruficaudus) chipmunks form a sister clade to the six T. quadrivittatus group species. Within this clade, T. canipes and T. rufus are each recovered as monophyletic (posterior probability = 1). The other four species (T. umbrinus, T. cinereicollis, T. dorsalis, and T. quadrivittatus) are dispersed throughout the tree with high posterior support. Because species were assigned using morphology, the compete mtDNA genome tree is in agreement with other studies (Good et al. 2003; Sullivan et al. 2014) in indicating extensive mitochondrial introgression.
Fig. 2.—
Maximum clade credibility tree estimated for the T. quadrivittatus group using mitochondrial genomes. Each split with a posterior probability greater than 0.9 is annotated. Colors correspond to species assignments. Representatives of T. amoenus, T. ruficaudus, and T. striatus are also included.
Shimodaira–Hasegawa tests reveal that each gene tree is not significantly different from the combined tree (supplementary table S3, Supplementary Material online). These results support the interpretation that mitochondrial genomes can be treated as a single marker due to linkage. Because a single underlying topology is supported among all loci, using the complete genome for phylogenetic inference is appropriate. It also confirms that previous studies that used cytochrome b or cytochrome oxidase subunit II as the sole marker should have recovered the correct underlying mitochondrial tree. This provides support for previous, single-locus studies in chipmunks.
Selection Analyses
Likelihood-ratio tests are not significant for any of the three comparisons (M1a vs. M2a; M7 vs. M8; M8 vs. M8a, supplementary table S1B, Supplementary Material online), with or without T. striatus. Estimating the posterior probability of each model is illuminating (supplementary table S1A, Supplementary Material online). High posterior probabilities are assigned to M5, and M7, models that do not include a site class where ω > 1. The T. dorsalis dataset follows the same pattern, as expected given that T. dorsalis shares mitochondrial haplotypes with other co-occurring T. quadrivittauts group species. Investigating model fit by traditional likelihood-ratio tests in concert with comparing posterior probabilities among models and model averaging of parameters allows for a more confident inference of patterns of selection. This approach could be applied in other studies where the posterior probabilities of a particular site being under selection can be inferred with greater confidence. Additionally, the branch test for differential selection within lineages of T. dorsalis fails to reject the null model of a single ω across the tree (P > 0.87, supplementary table S1B, Supplementary Material online), suggesting no lineage-specific differences in ω across T. dorsalis.
Pairwise McDonald–Kreitman tests reveal no significant deviations from neutrality after correcting for multiple tests (table 4). Tajima’s D, whether using all sites or a subset of sites, fails to detect any statistically significant deviations from neutrality (table 5). Our HyPhy selection analyses (SLAC, FEL, IFEL, REL, and FUBAR) detected almost no sites under positive or diversifying selection across all genes (table 6). In a handful of cases, a single codon of interest was identified. Examining the data revealed that this was a rare substitution present in a single sample, T. quadrivittatus, that was not present in other T. quadrivittatus samples. Based on this evidence, we conclude that no sites can be identified as being under positive or diversifying selection in this dataset.
Table 4.
McDonald–Kreitman Tests
| Species 1 | Species 2 | P-value | Interpretation (P < 0.003) |
|---|---|---|---|
| T. quadrivittatus | T. cinereicollis | N/A | Non-significant |
| T. quadrivittatus | T. rufus | 0.309 | Non-significant |
| T. quadrivittatus | T. canipes | 0.493 | Non-significant |
| T. quadrivittatus | T. umbrinus | 1 | Non-significant |
| T. cinereicollis | T. rufus | 0.059 | Non-significant |
| T. cinereicollis | T. canipes | 0.013 | Non-significant |
| T. cinereicollis | T. umbrinus | 0.498 | Non-significant |
| T. rufus | T. canipes | 0.005 | Non-significant |
| T. rufus | T. umbrinus | 0.327 | Non-significant |
| T. canipes | T. umbrinus | 0.022 | Non-significant |
| T. quadrivittatus | T. amoenus | 0.338 | Non-significant |
| T. cinereicollis | T. amoenus | 0.713 | Non-significant |
| T. umbrinus | T. amoenus | 0.423 | Non-significant |
| T. rufus | T. amoenus | 0.711 | Non-significant |
| T. canipes | T. amoenus | 0.905 | Non-significant |
Note.—Each quadrivittatus group species (excluding T. dorsalis, see details in the text) is evaluated in a pairwise fashion. Additionally, each species is evaluated relative to T. amoenus. N/A refers to the inability to calculate a test statistic due to a lack of nonsynonymous substitutions. NS refers to nonsignificant P-values as calculated in Dnasp through permutation and corrected for multiple tests.
Table 5.
Tests for Deviations from Neutrality
| Species | Tajima's D | P-value | Tajima's D (NS sites) | P-value | Tajima's D (S sites) | P-value |
|---|---|---|---|---|---|---|
| T. canipes | −0.98771 | Non-significant | −1.0154 | Non-significant | −0.9604 | Non-significant |
| T. cinereicollis | 0.07405 | Non-significant | −0.4992 | Non-significant | 0.148 | Non-significant |
| T. quadrivittatus | 0.95892 | Non-significant | 0.5447 | Non-significant | 1.0159 | Non-significant |
| T. rufus | 0.86489 | Non-significant | 1.7183 | Non-significant | 0.6202 | Non-significant |
| T. umbrinus | −0.0425 | Non-significant | −0.709 | Non-significant | −0.1688 | Non-significant |
Note:—Tajima’s D is estimated for all protein-coding sites, just nonsynonymous (NS) sites, and just synonymous (S) sites. Nonsignificant P-values are listed as ‘Non-significant’.
Table 6.
Model Averaged Parameter Estimates and Additional Tests for Positive Selection
| Model-averaged ω | Model-averaged κ | SLAC+ | FEL+ | IFEL+ | REL+ | FUBAR Diversifying | |
|---|---|---|---|---|---|---|---|
| Striatus | 0.022 | 9.877 | 0 | 1 | 1 | 0 | 0 |
| No striatus | 0.027 | 17.655 | 0 | 1 | 0 | 0 | 1 |
Note.—Estimates of ω and κ were obtained using a model-averaging approach over a series of models in PAML. Other selection analyses (SLAC, FEL, IFEL, REL, and FUBAR) were performed in HyPhy using the same dataset as the PAML selection analyses. Results are shown with and without T. striatus, the outgroup. Values represent the number of sites identified by each method as being under positive (or diversifying) selection.
Discussion
Mitochondrial introgression is often described in natural systems, and the relatively high frequency at which it occurs is surprising (Funk and Omland 2003). Because mitochondrial genomes contain protein-coding genes that act as part of the oxidative-phosphorylation pathway, as well as rRNAs that bind with ribosomal proteins encoded in the nuclear genome (and are imported into the mitochondria) for proper ribosomal assembly, it is reasonable to assume that antagonistic epistatic interactions with nuclear-encoded proteins from divergent lineages could reduce fitness (Ballard and Whitlock 2004; Ballard and Rand 2005). This has been described in several studies in Drosophila (Clark and Lyckegaard 1988; James and Ballard 2003). Selection, then, ought to act against introgression of divergent mtGenomes that introduce deleterious mitonuclear interactions (reviewed in Dowling et al. 2008).
The population genetic dynamics of mtDNA depend on relative fitness in concert with demographic factors. Haplotypes that confer high fitness may rise to a high frequency in a population following a selective sweep (Haldane 1924; Barton 2000). Alternatively, genotypes may invade resident populations through demographic means such as population expansion, and theory has shown that this results in asymmetric introgression of alleles from the resident population into the invading population (Excoffier et al. 2009). Furthermore, alleles at the front of an advancing population may also become fixed due to drift (Excoffier and Ray 2008; Hallatschek and Nelson 2008). Because these effects are at least partly stochastic, it may be impossible to predict the resulting dynamics a priori.
We did not detect positive selection acting on mitochondrial protein-coding genes in this group of chipmunks. Model-averaged estimates of ω are low (supplementary table S1C, Supplementary Material online). McDonald–Kreitman tests, Tajima’s D, and other selection tests implemented in HyPhy (SLAC, FEL, IFEL, REL, and FUBAR) detect no significant signatures of deviations from neutrality or positive selection. Neutrality indices are larger than one (data not shown), indicating an excess of nonsynonymous polymorphism and selection against slightly deleterious variation via purifying selection as described previously at mitochondrial loci (Nachman et al. 1994; Nachman 1998). However, it could be the case that positive selection does act on the mitochondrial genome, just not on the 13 protein-coding genes (Melo-Ferreira et al. 2014). Ribosomal RNAs interact with a number of nuclear proteins to construct a functional ribosome, suggesting that selection could act on these genes. In addition, the control region contains several promoter regions and other motifs, any of which could viably be targets for positive selection. Additionally, mitochondrial introgression could be facilitated by nuclear introgression (cytonuclear cointrogression; Beck et al. 2015), though recent work in chipmunks suggests little nuclear introgression even in cases where mitochondrial introgression is rampant (Good et al. 2015).
Nevertheless, in the absence of evidence for positive selection, we tentatively accept demographic factors, specifically population expansion, as a likely explanation that governs the extensive introgression of mtDNA in this system. T. dorsalis provides support for this conclusion. This species is broadly distributed in western North America, and its range overlaps with several other species. Interestingly, whenever it co-occurs with another species, it carries a mitochondrial genome that has introgressed from its congener. It is very likely that, during Pleistocene glacial maxima, T. dorsalis expanded to contact populations of other T. quadrivittatus group species (i.e., invaded existing populations) and, as a result, co-opted those species’ mitochondrial genomes (Currat et al. 2008). Niche modeling for T. dorsalis has been hind-casted onto climate reconstruction for the last glacial maximum by Waltari and Guralnick (2009), revealing that T. dorsalis has undergone a northward range expansion since the last glacial maximum, resulting in recent contact with other species. Currently, T. dorsalis-specific mtDNA is only found in southern refugial areas where no other species of Tamias occur (Sullivan et al. 2014).
However, the dynamics of the northern Rocky Mountains introgression of T. r. ruficaudus into T. a . luteiventris (Good et al. 2003) are less clear. The location of this introgression (along the crest of the Canadian Rockies) did not support forested habitats until just a few thousand years ago (Mack et al. 1978). Therefore, T. a. luteiventris has been undergoing population expansion in a similar fashion to T. r. ruficaudus, and differential rates of expansion could produce the observed pattern of introgression. A complete characterization of the dynamics of this introgressive event will require genome sequencing from several individuals within and around this contact zone.
It may also be the case that we cannot accurately estimate ω with a dataset of this size and across such recently diverged genomes (∼2.5 Ma; Sullivan et al. 2014). These analyses are likely influenced by the length of the sequence, the number of individuals, and the amount of sequence divergence among individuals (Anisimova et al. 2001, 2002; Wong et al. 2004). Thus, it may not be possible to estimate ω confidently with modestly divergent sequences relative to analyses of taxa that span deeper divergences. However, restricting our analyses to a single individual of each species removes interspecific polymorphism that might be misinterpreted as fixed variation, increasing the power of our analyses to accurately detect sites under positive selection.
Additionally, we develop a model-averaging approach that can be used to investigate patterns of selection in genomic-scale datasets. Estimating posterior probabilities for a range of models provides more information than discriminating among models using δAIC or likelihood-ratio tests alone. Furthermore, posterior probabilities can be used to weight parameters of interest, such as κ or ω, estimated under several models and mitigate biases that may be introduced from using a single model. We suggest use of the model-averaging approach described above to explicitly account for uncertainty in model selection, especially with regard to detecting positive selection when using likelihood-based approaches.
In conclusion, we sequence and characterize mitochondrial genomes for several species of chipmunks. Protein-coding genes are analyzed and used to test demographic vs. selection hypotheses governing introgression. We find no evidence for positive selection at mitochondrial loci and conclude that introgression is mediated by demographic factors (i.e., population expansion) in this system. Future analyses will focus on quantifying the amount of nuclear introgression taking place in this complex system, building on the characterizations in this study.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors thank the following for assistance in the field over several years: K. Bell, W. Bell, I. Demboski, M. Fraker, D. Good, P. Good, J. Harper, A. Hornsby, S. Poler, A. Runck, and D. Sullivan. We thank B. Martin, J. Gonzalez, and four anonymous reviewers for helpful comments that improved the manuscript. We also thank the Denver Museum of Nature & Science for providing tissue loans. This research was conducted in compliance with University of Idaho Animal Care and Use Committee, under protocol UIACUC-2005-40, and was supported by a seed grant from the University of Idaho Research Foundation, the NSF EPSCoR program (NSF cooperative agreement number EPS-9720634), the Institute for Bioinformatics and Evolutionary Studies (IBEST) at the University of Idaho (by NIH NCRR 1P20RR016454-01; NIH NCRR 1P20RR016448-01; NSF EPS-809935), NSF DEB-0717426 (JS), NSF DEB-0716200 (JRD), NSF Cooperative Agreement No. DBI-0939454 (JS), and the Denver Museum of Nature & Science. JS and BAJS received funding through BEACON, an NSF-funded Center the Study of Evolution in Action (DBI-0939454). Craig Moritz, Ke Bi, and Tyler Linderoth helped develop exon capture in chipmunks, supported by the University of California, Berkeley VCR-BiGCB and an NSERC Postdoctoral Fellowship to Bi. Additionally, this work was supported by start-up funds from the University of Montana (JMG) and the Gordon and Betty Moore Foundation. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
Literature Cited
- Abdo Z, Minin VN, Joyce P, Sullivan J. 2005. Accounting for uncertainty in the tree topology has little effect on the decision-theoretic approach to model selection in phylogeny estimation. Mol Biol Evol. 22:691–703. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Bielawski JP, Yang Z. 2002. Accuracy and power of Bayes prediction of amino acid sites under positive selection. Mol Biol Evol. 19:950–958. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Bielawski JP, Yang Z. 2001. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol Biol Evol. 18:1585–1592. [DOI] [PubMed] [Google Scholar]
- Axel J, Feldmaier-Fuchs G, Thomas WK, von Haeseler A, Paabo S. 1994. The marsupial mitochondrial genome and the evolution of placental mammals. Genetics 137:243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard JWO, Rand DM. 2005. The population biology of mitochondrial DNA and its phylogenetic implications. Annu Rev Ecol Evol Syst. 36:621–642. [Google Scholar]
- Ballard JWO, Whitlock MC. 2004. The incomplete natural history of mitochondria. Mol Ecol. 13:729–744. [DOI] [PubMed] [Google Scholar]
- Barton NH. 2000. Genetic hitchhiking. Philos Trans R Soc Lond B Biol Sci. 355:1553–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck EA, Thompson AC, Sharbrough J, Brud E, Llopart A. 2015. Gene flow between Drosophila yakuba and Drosophila santomea in subunit V of cytochrome c oxidase: a potential case of cytonuclear cointrogression. Evolution 69:1973–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm JE, Ives AR, Boughman JW. 2010. Breakdown in postmating isolation and the collapse of a species pair through hybridization. Am Nat. 175:11–26. [DOI] [PubMed] [Google Scholar]
- Best TL. 1993. Tamias ruficaudus. Mamm Species 1–7. [Google Scholar]
- Bi K, et al. 2012. Transcriptome-based exon capture enables highly cost-effective comparative genomic data collection at moderate evolutionary scales. BMC Genomics 13:403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossu CM, Near TJ. 2009. Gene trees reveal repeated instances of mitochondrial DNA introgression in orange throat darters (Percidae: Etheostoma). Syst Biol. 58:114–129. [DOI] [PubMed] [Google Scholar]
- Brown JH. 1971. Mechanisms of competitive exclusion between two species of chipmunks. Ecology 52:305–311. [Google Scholar]
- Callahan J. 1981. Vocal solicitation and parental investment in female Eutamias. 118:872–875. [Google Scholar]
- Chang JH, Tong L. 2012. Mitochondrial poly(A) polymerase and polyadenylation. Biochim Biophys Acta. 1819:992–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Lyckegaard EMS. 1988. Natural selection with nuclear and cytoplasmic transmission. III. Joint analysis of segregation and mtDNA in Drosophila melanogaster . Genetics 118:471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currat M, Excoffier L. 2004. Modern humans did not admix with Neanderthals during their range expansion into Europe. PLoS Biol. 2:e421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currat M, Ruedi M, Petit RJ, Excoffier L. 2008. The hidden side of invasions: massive introgression by local genes. Evolution 62:1908–1920. [DOI] [PubMed] [Google Scholar]
- Dorner M, Altmann M, Paabo S, Morl M. 2001. Evidence for import of a lysyl-tRNA into marsupial mitochondria. Mol Biol Cell 12:2688–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling DK, Friberg U, Lindell J. 2008. Evolutionary implications of non-neutral mitochondrial genetic variation. Trends Ecol Evol. 23:546–554. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 29:1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds CA, Lillie AS, Cavalli-Sforza LL. 2004. Mutations arising in the wave front of an expanding population. Proc Natl Acad Sci U S A. 101:975–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans J, Sullivan J. 2011. Approximating model probabilities in Bayesian information criterion and decision-theoretic approaches to model selection in phylogenetics. Mol Biol Evol. 28:343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Foll M, Petit RJ. 2009. Genetic consequences of range expansions. Annu Rev Ecol Evol Syst. 40:481–501. [Google Scholar]
- Excoffier L, Ray N. 2008. Surfing during population expansions promotes genetic revolutions and structuration. Trends Ecol Evol. 23:347–351. [DOI] [PubMed] [Google Scholar]
- Funk DJ, Omland KE. 2003. Species-level paraphyly and polyphyly: frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annu Rev Ecol Evol Syst. 34:397–423. [Google Scholar]
- Gambs RD. 1965. Zoogeography of Chipmunks in the Pacific Northwest: A Thesis. University of Idaho, Moscow.
- Gilman RT, Behm JE. 2011. Hybridization, species collapse, and species reemergence after disturbance to premating mechanisms of reproductive isolation. Evolution 65:2592–2605. [DOI] [PubMed] [Google Scholar]
- Good JM, et al. 2008. Ancient hybridization and mitochondrial capture between two species of chipmunks. Mol Ecol. 17:1313–1327. [DOI] [PubMed] [Google Scholar]
- Good JM, Demboski JR, Nagorsen DW, Sullivan J. 2003. Phylogeography and introgressive hybridization: chipmunks (genus Tamias) in the northern Rocky Mountains. Evolution 57:1900–1916. [DOI] [PubMed] [Google Scholar]
- Good JM, Sullivan J. 2001. Phylogeography of the red-tailed chipmunk (Tamias ruficaudus), a northern Rocky Mountain endemic. Mol Ecol. 10:2683–2695. [DOI] [PubMed] [Google Scholar]
- Good JM, Vanderpool D, Keeble S, Bi K. 2015. Negligible nuclear introgression despite complete mitochondrial capture between two species of chipmunks. Evolution 69:1961–1972. [DOI] [PubMed] [Google Scholar]
- Gray AP. 1972. Mammalian hybrids: a check-list with bibliography. 2nd ed. Slough: Commonwealth Agricultural Bureau. [Google Scholar]
- Haldane JBS. 1924. A mathematical theory of natural and artificial selection—I. Trans. Cambridge Philos Soc. 23:19–41. [Google Scholar]
- Hallatschek O, Hersen P, Ramanathan S, Nelson DR. 2007. Genetic drift at expanding frontiers promotes gene segregation. Proc Natl Acad Sci U S A. 104:19926–19930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallatschek O, Nelson DR. 2008. Gene surfing in expanding populations. Theor Popul Biol. 73:158–170. [DOI] [PubMed] [Google Scholar]
- Hedrick PW. 2013. Adaptive introgression in animals: examples and comparison to new mutation and standing variation as sources of adaptive variation. Mol Ecol. 22:4606–4618. [DOI] [PubMed] [Google Scholar]
- Heller HC. 1971. Altitudinal zonation of chipmunks (Eutamias): interspecific aggression. Ecology 52:312. [Google Scholar]
- Hird S, Reid N, Demboski J, Sullivan J. 2010. Introgression at differentially aged hybrid zones in red-tailed chipmunks. Genetica 138:869–883. [DOI] [PubMed] [Google Scholar]
- Hird S, Sullivan J. 2009. Assessment of gene flow across a hybrid zone in red-tailed chipmunks (Tamias ruficaudus). Mol Ecol. 18:3097–3109. [DOI] [PubMed] [Google Scholar]
- Hofacker IL. 2003. Vienna RNA secondary structure server. Nucleic Acids Res. 31:3429–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter SS, et al. 2015. Assembly by Reduced Complexity (ARC): a hybrid approach for targeted assembly of homologous sequences. bioRxiv. http://biorxiv.org/lookup/doi/10.1101/014662
- James AC, Ballard JWO. 2003. Mitochondrial genotype affects fitness in Drosophila simulans . Genetics 164:187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanet A, Secondi J, Lemaire C. 2011. Widespread introgression does not leak into allotopy in a broad sympatric zone. Heredity 106:962–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahle D, Wickham H. 2013. ggmap : spatial visualization with ggplot2. R J. 5:144–161. [Google Scholar]
- Katoh K, Toh H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 9:286–298. [DOI] [PubMed] [Google Scholar]
- Kearse M, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfstein S, Currat M, Excoffier L. 2006. The fate of mutations surfing on the wave of a range expansion. Mol Biol Evol. 23:482–490. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, et al. 2006. Adaptation to different human populations by HIV-1 revealed by codon-based analyses. PLoS Comput Biol. 2:e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SDW. 2005a. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531–2533. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SDW. 2005b. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 22:1208–1222. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SDW, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. [DOI] [PubMed] [Google Scholar]
- Llopart A, Herrig D, Brud E, Stecklein Z. 2014. Sequential adaptive introgression of the mitochondrial genome in Drosophila yakuba and Drosophila santomea . Mol Ecol. 23:1124–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo A, et al. 2010. Performance of criteria for selecting evolutionary models in phylogenetics: a comprehensive study based on simulated datasets. BMC Evol Biol. 10:242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack RN, Rutter NW, Bryant VM, Valastro S. 1978. Reexamination of postglacial vegetation history in northern Idaho: Hager Pond, Bonner Co. Quat Res. 10:241–255. [Google Scholar]
- Magoč T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet J. 2005. Hybridization as an invasion of the genome. Trends Ecol Evol. 20:229–237. [DOI] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. 1991. Adaptive protein evolution at the Adh locus in Drosophila . Nature 351:652–654. [DOI] [PubMed] [Google Scholar]
- McGuire JA, et al. 2007. Mitochondrial introgression and incomplete lineage sorting through space and time: phylogenetics of crotaphytid lizards. Evolution 61:2879–2897. [DOI] [PubMed] [Google Scholar]
- Melo-Ferreira J, et al. 2014. The elusive nature of adaptive mitochondrial DNA evolution of an arctic lineage prone to frequent introgression. Genome Biol Evol. 6:886–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minin V, Abdo Z, Joyce P, Sullivan J. 2003. Performance-based selection of likelihood models for phylogeny estimation. Syst Biol. 52:674–683. [DOI] [PubMed] [Google Scholar]
- Murrell B, et al. 2013. FUBAR: a fast, unconstrained bayesian approximation for inferring selection. Mol Biol Evol. 30:1196–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman MW. 1998. Deleterious mutations in animal mitochondrial DNA. Genetica 102–103:61–69. [PubMed] [Google Scholar]
- Nachman MW, Boyer SN, Aquadro CF. 1994. Nonneutral evolution at the mitochondrial NADH dehydrogenase subunit 3 gene in mice. Proc Natl Acad Sci U S A. 91:6364–6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson BD, Norris RW. 2016. Towards a uniform nomenclature for ground squirrels: the status of the Holarctic chipmunks. Mammalia 80: [Google Scholar]
- Petit RJ, Excoffier L. 2009. Gene flow and species delimitation. Trends Ecol Evol. 24:386–393. [DOI] [PubMed] [Google Scholar]
- Piaggio AJ, Spicer GS. 2000. Molecular phylogeny of the chipmunk genus Tamias based on the Mitochondrial cytochrome oxidase subunit II gene. J Mamm Evol. 7:147–166. [Google Scholar]
- Piaggio AJ, Spicer GS. 2001. Molecular phylogeny of the chipmunks inferred from mitochondrial cytochrome b and cytochrome oxidase II gene sequences. Mol Phylogenet Evol. 20:335–350. [DOI] [PubMed] [Google Scholar]
- Pinho C, Hey J. 2010. Divergence with gene flow: models and data. Annu Rev Ecol Evol Syst. 41:215–230. [Google Scholar]
- Posada D, Buckley TR. 2004. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol. 53:793–808. [DOI] [PubMed] [Google Scholar]
- Raftery AE. 1995. Bayesian model selection in social research. Sociol Methodol. 25:111. [Google Scholar]
- Reid N, Demboski JR, Sullivan J. 2012. Phylogeny estimation of the radiation of western North American chipmunks (Tamias) in the face of introgression using reproductive protein genes. Syst Biol. 61:44–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid N, Hird S, Schulte-Hostedde A, Sullivan J. 2010. Examination of nuclear loci across a zone of mitochondrial introgression between Tamias ruficaudus and T. amoenus . J Mammal. 91:1389–1400. [Google Scholar]
- Reyes A, Gissi C, Pesole G, Catzeflis FM, Saccone C. 2000. Where do rodents fit? Evidence from the complete mitochondrial genome of Sciurus vulgaris . Mol Biol Evol. 17:979–983. [DOI] [PubMed] [Google Scholar]
- Reyes A, Pesole G, Saccone C. 1998. Complete mitochondrial DNA sequence of the fat dormouse, Glis glis: further evidence of rodent paraphyly. Mol Biol Evol. 15:499–505. [DOI] [PubMed] [Google Scholar]
- Rheindt FE, Edwards SV. 2011. Genetic introgression: an integral but neglected component of speciation in birds. Auk 128:620–632. [Google Scholar]
- Rice WR, Hostert EE. 1993. Laboratory experiments on speciation: what have we learned in 40 years? Evolution 47:1637. [DOI] [PubMed] [Google Scholar]
- Ryan ME, Johnson JR, Fitzpatrick BM. 2009. Invasive hybrid tiger salamander genotypes impact native amphibians. Proc Natl Acad Sci U S A. 106:11166–11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte-Hostedde AI, Millar JS. 2004. Intraspecific variation of testis size and sperm length in the yellow-pine chipmunk (Tamias amoenus): implications for sperm competition and reproductive success. Behav Ecol Sociobiol. 55:272–277. [Google Scholar]
- Schulte-Hostedde AI, Millar JS, Gibs HL. 2004. Sexual selection and mating patterns in a mammal with female-biased sexual size dimorphism. Behav Ecol. 15:351–356. [Google Scholar]
- Schwarz G. 1978. Estimating the dimension of a model. Ann Stat. 6:461–464. [Google Scholar]
- Shimodaira H, Hasegawa M. 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 16:1114–1116. [Google Scholar]
- Sullivan J, et al. 2014. Divergence with gene flow within the recent chipmunk radiation (Tamias). Heredity 113:185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan J, Joyce P. 2005. Model selection in phylogenetics. Annu Rev Ecol Evol Syst. 36:445–466. [Google Scholar]
- Swofford DL. 2003. Phylogenetic Analysis Using Parsimony (*and Other Methods). Sunderland (MA: ): Sinauer Associates. [Google Scholar]
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor EB, et al. 2006. Speciation in reverse: morphological and genetic evidence of the collapse of a three-spined stickleback (Gasterosteus aculeatus) species pair. Mol Ecol. 15:343–355. [DOI] [PubMed] [Google Scholar]
- Thorington Jr, RW, Hoffman RS. Family Sciuridae In: Wilson DE, Reeder DM, editors. Mammal species of the world: a taxonomic and geographic reference.. Baltimore (MD: ): Johns Hopkins University Press, p. 754–818. [Google Scholar]
- Travis JMJ, et al. 2007. Deleterious mutations can surf to high densities on the wave front of an expanding population. Mol Biol Evol. 24:2334–2343. [DOI] [PubMed] [Google Scholar]
- Untergasser A, et al. 2007. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35:W71–W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltari E, Guralnick RP. 2009. Ecological niche modelling of montane mammals in the Great Basin, North America: examining past and present connectivity of species across basins and ranges. J Biogeogr. 36:148–161. [Google Scholar]
- Wong WSW, Yang Z, Goldman N, Nielsen R. 2004. Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics 168:1041–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-I. 2001. The genic view of the process of speciation. J Evol Biol. 14:851–865. [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Zwickl DJ. 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Austin (TX: ): The University of Texas at Austin. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.