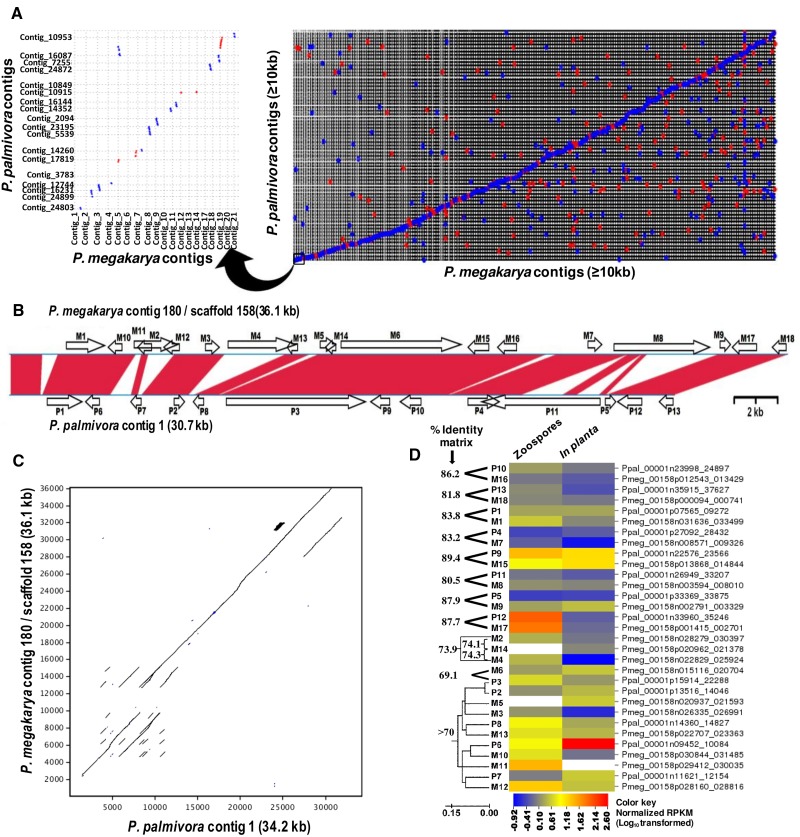
Fig. 2.—
Synteny- and repeat-driven genome expansion between the P. megakarya and P. palmivora genomes. (A) MUMmer alignment dot plot of P. megakarya and P. palmivora contigs (≥10 kb) (NUCmer parameters: breaklen = 200, maxgap = 90, mincluster = 65, minmatch = 20). Red dots represent positive strand alignments whereas blue dots represent negative strand alignments. The left panel is an enlargement of the bottom-left corner of the right panel. (B) Conserved gene order and genome expansion across two homologous contigs of the P. megakarya and P. palmivora genomes. Phytophthora megakarya contig 180 and P. palmivora contig 1 were aligned using Lasergene’s MegAlign Pro and gene model locations (indicated by block arrows) were determined manually. Red bands represent homologous regions (≥65% nucleotide sequence identity in either orientation). (C) LALIGN pairwise sequence alignment (Huang and Miller 1991) of the two contigs showing repeat-driven expansion in three different regions of the contigs, in addition to a 2.3-kb insertion in the P. megakarya contig. (D) Sequence similarities among the PGeneMs within the two contigs and their expression profiles as determined by RNA-Seq analysis. Amino acid identities between closely related PGeneMs (>70% identity) were inferred using guide trees generated by WebPRANK (http://www.ebi.ac.uk/goldman-srv/webprank/, last accessed 16 February 2017). Branch lengths in the tree represent the number of nucleic acid substitutions per site. RNA-Seq read counts for zoospore and in planta libraries were normalized by read counts in the mycelium library and values were LOG-transformed to linearize the data. The heat map was generated using CIMminer (http://discover.nci.nih.gov/cimminer, last accessed 16 February 2017). Underlying % Identity Matrix data are shown in the supplementary Excel file S1, sheet 7: %IM Pmeg-Ppal contig, Supplementary Material online.