Abstract
Unlike hexapods and vertebrates, in chelicerates, knowledge of the specific molecules involved in chemoreception comes exclusively from the comparative analysis of genome sequences. Indeed, the genomes of mites, ticks and spiders contain several genes encoding homologs of some insect membrane receptors and small soluble chemosensory proteins. Here, we conducted for the first time a comprehensive comparative RNA-Seq analysis across different body structures of a chelicerate: the nocturnal wandering hunter spider Dysdera silvatica Schmidt 1981. Specifically, we obtained the complete transcriptome of this species as well as the specific expression profile in the first pair of legs and the palps, which are thought to be the specific olfactory appendages in spiders, and in the remaining legs, which also have hairs that have been morphologically identified as chemosensory. We identified several ionotropic (Ir) and gustatory (Gr) receptor family members exclusively or differentially expressed across transcriptomes, some exhibiting a distinctive pattern in the putative olfactory appendages. Furthermore, these IRs were the only known olfactory receptors identified in such structures. These results, integrated with an extensive phylogenetic analysis across arthropods, uncover a specialization of the chemosensory gene repertoire across the body of D. silvatica and suggest that some IRs likely mediate olfactory signaling in chelicerates. Noticeably, we detected the expression of a gene family distantly related to insect odorant-binding proteins (OBPs), suggesting that this gene family is more ancient than previously believed, as well as the expression of an uncharacterized gene family encoding small globular secreted proteins, which appears to be a good chemosensory gene family candidate.
Keywords: chemosensory gene families, specific RNA-Seq, de novo transcriptome assembly, functional annotation, chelicerates, arthropods
Introduction
Chemoreception, the detection and processing of chemical signals in the environment, is a biological process that is critical for animal survival and reproduction. The essential role of smell and taste in the detection of food, hosts and predators and their participation in social communication make the molecular components of this system solid candidates for important adaptive changes associated with animal terrestrialization (Whiteman and Pierce 2008). In insects, chemical recognition occurs in specialized hair-like cuticular structures called sensilla, which can be found almost anywhere in the body (Joseph and Carlson 2015). In Drosophila, olfactory sensilla are concentrated on the antenna and the maxillary palps, while gustatory sensilla are spread across various body locations, such as the proboscis, the legs and the anterior margins of wings (Pelosi 1996; Shanbhag et al. 2001). The chemoreceptor proteins embedded within the membrane of sensory neurons (SN) innervating these sensilla are responsible for transducing the external chemical signal into an action potential. In the case of smell, olfactory SNs project the axons to specific centers of the brain, where the signals are processed and engender a behavioral response to the specific external stimuli. The process can be facilitated by small soluble chemosensory proteins that are secreted in the lymph that bathes the dendrites of the SNs and are believed to solubilize and either transport the signaling molecules to membrane receptors or protect them from premature degradation (Vogt and Riddiford 1981; Pelosi et al. 2006). Although insect chemoreceptors and soluble chemosensory proteins are encoded by gene families exhibiting high gene turnover rates (see Sánchez-Gracia et al. 2011 for a comprehensive review), distant homologues of the members of these families have been identified in other arthropod lineages (Colbourne et al. 2011; Vieira and Rozas 2011; Chipman et al. 2014; Frías-López et al. 2015; Gulia-Nuss et al. 2016). Vertebrate functional counterparts of these gene families, however, are not evolutionarily related; indeed, the members of this subphylum use different molecules to perform the same general physiological function (Kaupp 2010).
Spiders comprise a highly diverse group of arthropods, including >45,000 described species (World Spider Catalog 2016), and are dominant predators in most terrestrial ecosystems. Given their potential as biological control agents as well as the engineering properties of silk and venom, these organisms are of great economic and medical relevance (Clarke et al. 2014). Because the Arachnida ancestors of these chelicerates colonized the land ∼475 Ma, long after the split of the four major extant arthropod lineages (Rota-Stabelli et al. 2013), spiders are good models for comparative studies on the diverse strategies adopted by arthropod lineages during their independent adaptation to terrestrial environments. However, despite their biological and translational implications, there are relatively few genomic and transcriptomic studies conducted on these organisms compared with those conducted on insects, and studies on spiders almost exclusively focus on silk and venom research (Grbić et al. 2011; Clarke et al. 2014; Posnien et al. 2014; Sanggaard et al. 2014).
Spiders can detect volatile and nonvolatile compounds through specialized chemosensitive hairs distributed at the tips of various extremities and appendages, including legs and palps (Foelix 1970; Foelix and Chu-Wang 1973; Kronestedt 1979; Cerveira and Jackson 2012; Foelix et al. 2012). Nevertheless, the molecular nature of chelicerate chemoreceptors has remained elusive until recently. We and others have identified distant homologs of some insect gene families associated with chemosensation in the genomes of mites, ticks and spiders (Montagné et al. 2015; Gulia-Nuss et al. 2016), such as members of the gustatory (Gr) and ionotropic (Ir) receptor, and of the chemosensory protein (Csp), Niemann–Pick protein type C2 (Npc2) and sensory neuron membrane protein (Snmp) multigene families. In addition, chelicerates lack homologs of the typical insect olfactory receptor family Ors, which are thought to have originated later with the appearance of flying insects, and no Obp gene had been detected to date (Vieira and Rozas 2011; Chipman et al. 2014). Overall, available genomic studies suggest that the Ir gene family is responsible for smell not only in chelicerates but also in all nonneopteran arthropods (Croset et al. 2010; Colbourne et al. 2011; Chipman et al. 2014; Gulia-Nuss et al. 2016). Regarding taste, the presence of numerous copies of Gr and nonconserved Ir (a group of divergent IR proteins associated with gustatory function in insects, Croset et al. 2010) genes in chelicerate genomes clearly suggests that these families are responsible for contact chemoreception in this species.
Nevertheless, the simple comparative analysis of genomic sequences does not allow inferring which specific members of already known chemosensory families are involved in the different sensory modalities. Additionally, chelicerates could also use molecules completely different from those already known in insects during the water-to-land transition, which should also be different from those used by vertebrates (these molecules have also not been found in the available genome sequences); these uncharacterized genes (or annotated with incomplete gene models) would be not directly detectable only by comparative genomics. Instead, specific transcriptomic analyses of chemosensory tissues can provide useful insight into all these issues. Antennae-specific gene expression studies in lobsters and hermit crabs (Corey et al. 2013; Groh-Lunow et al. 2014), for example, have revealed the presence of several transcripts encoding IRs, supporting the active role in olfaction of this gene family in crustaceans. To gain insight into the specific proteins involved in chelicerate chemoreception, we recently performed a tissue-specific comparative transcriptomics study in the funnel-web spider Macrothele calpeiana (Frías-López et al. 2015). Unfortunately, we failed to detect the specific expression of Ir or Gr genes in the first pair of legs and in palps, the best candidate structures to hold olfactory hairs in chelicerates. This result might be caused by either the sedentary lifestyle of this mygalomorph spider, which may lead to a marginal role of chemical communication in this species, or the low sequencing coverage of this RNA-Seq study.
Here, in order to better characterize the chemosensory repertoire of a spider, we report a more comprehensive comparative transcriptomic analysis in an active nocturnal hunter spider, Dysdera silvatica Schmidt, 1981 (Araneae, Dysderidae) (fig. 1). This species, which is endemic to the Canary Islands, belongs to a genus characterized by long and protruding chelicerae used to capture and feed on woodlice (Crustacea: Isopoda: Oniscidea; fig. 1B ). We have conducted a deep RNA-Seq experiment in four separated body parts, three of them likely containing chemosensitive hairs in spiders. Because the performance of the de novo assembly of short reads strongly depends on biological data (i.e., the complexity of the data is almost species specific), we first performed a comparative analysis among a set of commonly used software for transcriptome assembly. Based on the best assembly and highly accurate functional annotations, we conducted a comparative analysis between the specific transcriptomes of the different body parts, emphasizing the detection of distinctive chemosensory profiles, especially in the palps and the first pair of legs, which has been reported to hold the peripheral olfactory structures in spiders. We then contextualized these results by applying a sound phylogenetic analysis including representative members of each arthropod chemosensory gene family.
Fig. 1.—
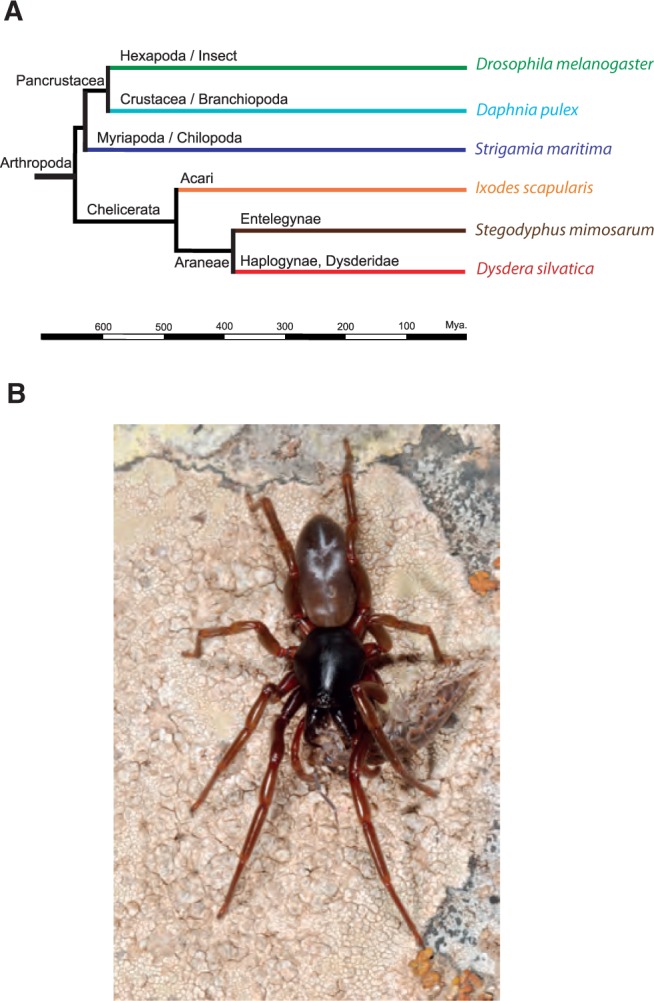
(A) Phylogenetic position of Dysdera silvatica within arthropods. Divergence times were obtained from TimeTree (Hedges et al. 2015). (B) D. silvatica feeding on a woodlouse.
We have identified several members of the Ir and Gr gene families specifically or differently expressed in some of the four surveyed transcriptomes (including a clear homolog of the co-receptor IR25a of Drosophila melanogaster) and some signs of chemosensory specialization across spider chemosensory structures. Moreover, we have also identified three genes distantly related to the insect Obp gene family and a new gene family encoding small secreted soluble proteins that might function as molecular carriers in the spider chemosensory system. We discuss these findings in the context of the origin and evolution of chemosensory gene families in arthropods and propose some candidate genes that may have an important chemoreceptor role in spiders.
Materials and Methods
Sample Collection, RNA Extraction and Library Preparation
We sequenced and analysed the transcriptome of four D. silvatica males (voucher specimens were deposited at the Centre de Recursos de Biodiversitat Animal of the Universitat de Barcelona under catalog numbers NMH2597-99 and NMH2601) collected from the Canary Islands, La Gomera and Las Tajoras (28.112736 N, 17.262511 W) in 2013. We used males because this sex has been shown to respond to sex-specific olfactory information (Nelson et al. 2012). We performed four separated RNA-Seq experiments, which included expressed sequences form the palps (PALP), the first pair of legs (LEG#1), all other pairs of legs (LEG#234) and the remaining body structures (REST), henceforth referred to as experimental conditions. We dissected these body parts independently for each of the four males (after snap freezing in liquid nitrogen) and extracted the total RNA separately for each condition and sample using the RNeasy Mini kit (Qiagen, Venlo, The Netherlands) and TRIzol reagent (Invitrogen, Waltham, MA). We determined the amount and integrity of RNA using a Qubit Fluorometer (Life Technologies, Grand Island, NY) and Agilent 2100 Bioanalyzer (CCiTUB, Barcelona, Spain), respectively. We sequenced the transcriptome of each condition using the Illumina Genome Analyzer HiSeq 2000 (100 bp PE reads) according to the manufacturer’s instructions (Illumina, San Diego, CA). Briefly, for each experimental condition, the mRNA was purified from 1 µg of total RNA using magnetic oligo(dT) beads and fragmented into small pieces. Double-stranded cDNA was synthesized with random hexamer (N6) primers (Illumina), and Illumina paired-end (PE) adapters were ligated to the ends of adenylated cDNA fragments. All library preparation steps and transcriptome sequencing were carried out in Macrogen Inc., Seoul, South Korea.
Raw Data Pre-Processing
Raw NGS data were pre-processed to eliminate all reads with a quality score ≤20 in at least the 30% of the read length and to remove reads with putative sequencing errors using NGSQCToolkit and SEECER v_0.1.3 (Patel and Jain 2012; Le et al. 2013). Before the assembly step, we performed an in silico normalization of filtered reads using Diginorm, an algorithm included in Trinity software (Haas et al. 2014). We set 50X as the targeted maximum coverage for the reads.
De N ovo Transcriptome Assembly
First, to determine the best assembler for the D. silvatica RNA-Seq data, we compared the performance of five commonly used software programs in assembling the specific transcriptome of the experimental condition REST. We tested Trinity r2.1.1, Bridger r2014-12-01, SOAPdenovo-Trans release 1.03, Oases version_0.2.8, and ABySS version_1.3.7/trans-ABySS version1.4.8 (Birol et al. 2009; Schulz et al. 2012; Xie et al. 2014; Z. Chang et al. 2015). For this comparative analysis and depending on the specificities of the selected software (allowing single or multiple k-mer values), we applied several single k-mer lengths and k-mer ranges (see supplementary table S1, Supplementary Material online, for details).
After the assembly phase, we removed all contigs with evidence of contaminant sequences using the software Seqclean (ftp://occams.dfci.harvard.edu/pub/bio/tgi/software/; last accessed May 1, 2015) together with the sequences of the UniVec vector database and the genomes of Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus, Saccharomyces cerevisiae and Homo sapiens. Clean contigs were then clustered into putative transcripts (analogous to the Trinity components). We determined the assembly performance of each software based on (1) the DETONATE score (Li et al. 2014), (2) the outcome of the assembled sequences in a set of sequence similarity and profile-based searches using different databases (see the “Results” section for more details), and (3) some commonly used descriptive statistics on assembly quality, namely the average sequence length, the N50, the maximum and minimum transcript lengths and the total bases in the assembly, calculated with the NGSQCToolkit software and some Perl scripts. All analyses were run in a 64-CPU machine with 750 Gb of RAM.
Protein Databases
We built two customized protein databases to assist the functional annotation of the D. silvatica transcriptome. The arthropodDB database contains the publicly available amino acid sequences of fully annotated proteins and protein models from a set of representative arthropod genomes and some appropriated external groups, along with their complete entry description, associated GO terms and InterPro identifiers (Ashburner et al. 2000; Mitchell et al. 2014). This database includes information for the following species: (1) the chelicerates Ixodes scapularis (Acari) (Gulia-Nuss et al. 2016), Metaseiulus occidentalis (Acari) ( https://www.hgsc.bcm.edu/arthropods/western-orchard-predatory-mite-genome-project; last accessed May 1, 2015), Tetranychus urticae (Acari) (Grbić et al. 2011), Mesobuthus martensii (Scorpiones) (Cao et al. 2013), Acanthoscurria geniculata (Araneae, Theraphosidae) (Sanggaard et al. 2014) , Stegodyphus mimosarum (Araneae, Eresidae) (Sanggaard et al. 2014) , Latrodectus hesperus (Araneae) (https://www.hgsc.bcm.edu/arthropods/western-black-widow-spider-genome-project; last accessed May 1, 2015), Loxosceles reclusa (Araneae, Sicariidae) (https://www.hgsc.bcm.edu/arthropods/brown-recluse-spider-genome-project; last accessed May 1, 2015) and Parasteatoda tepidariorum (Araneae, Theridiidae) (https://www.hgsc.bcm.edu/arthropods/common-house-spider-genome-project; last accessed May 1, 2015); (2) the hexapods D. melanogaster (Diptera) (Adams et al. 2000), Pediculus humanus (Phthiraptera) ( Kirkness et al. 2010) and Bombyx mori (Lepidoptera) (Mita et al. 2004) ; (3) the crustacean Daphnia pulex (Branchiopoda) (Colbourne et al. 2011); (4) the myriapod Strigamia maritima (Chilopoda, Geophilomorpha) (Chipman et al. 2014); (5) the tardigrade Hypsibius dujardini (http://badger.bio.ed.ac.uk/H_dujardini; last accessed May 1, 2015); and (6) the nematode Caenorhabditis elegans. In the cases where there was no functional description or associated GO term (e.g., the protein models from A. geniculata, L. hesperus, L. reclusa, M. martensii, M. occidentalis and P. tepidariorum), we approximated the functional annotation using InterProScan version 5.4.47 (Jones et al. 2014).
The chemDB database contains the amino acid sequences and the functional information of all well-annotated members of the Or, Gr Ir, Csp, Obp, Npc2 and Snmp gene families from a representative set of insect species, namely D. melanogaster, Tribolium castaneum (Coleoptera), Apis mellifera (Hymenoptera) and Acyrthosiphon pisum (Hemiptera), and from the noninsect species included in arthropodDB. Moreover, we also included in chemDB some vertebrate odorant binding proteins and olfactory and taste receptors identified by the InterPro signatures IPR002448, IPR000725 and IPR007960, respectively (see supplementary table S1 B in Frías-López et al. 2015). Furthermore, we progressively updated chemDB by adding to this database all novel members of these chemosensory families (the conceptual translation of the identified transcripts) characterized in D. silvatica.
Functional Annotation of the D. s ilvatica Transcripts
We applied a similarity-based search approach to assist the annotation of the D. silvatica transcriptome. We first used BLASTx to search the translated transcripts against the SwissProt and arthropodDB databases (BLAST v2.2.29; Altschul et al. 1990; Altschul 1997). To search against NCBI-nr, we used GHOSTZ version1.0.0; this software is much faster than BLAST, especially for large databases without a substantial reduction of sensibility (Suzuki et al. 2014). We improved the functional annotation by searching for the specific protein-domain signatures in translated transcriptome sequences using InterProScan (Jones et al. 2014). We predicted signal peptides and transmembrane helices with SignalP and TMHMM, respectively (Krogh et al. 2001; Petersen et al. 2011). To carry out the profile-based searches, we created custom HMM models, one for each chemosensory family included in chemDB. These models are based on multiple sequence alignments (MSA) built with the program hmmalign (HMMER 3.1b1 package; Eddy 2011) using the specific core Pfam profile as a guide.
We conducted a GO-enrichment analysis with the BLAST2GO term suite using all functionally annotated transcripts with an associated GO term (Conesa et al. 2005). Moreover, we also searched these functionally annotated transcripts for KEGG enzymes and pathways (Kanehisa and Goto 2000), for CEG (Core Eukaryotic Genes) (Parra et al. 2007; Parra et al. 2009) and for the list of housekeeping (HK) genes used in supplementary table S1 A in Frías-López et al. (2015).
To characterize the chemosensory gene repertory of D. silvatica, we first used the proteins in chemDB as query sequences to search for putative homologs among spider transcripts (using tBLASTn search; E-value cutoff of 10 − 3). We only considered as positives those hits covering at least 2/3 of the query sequence length or the 80% of the total subject sequence. Then, we conducted some additional searches based on our custom HMM models and the conceptual translation of D. silvatica transcripts as subject sequences (using hmmer and an i-E-value of 10− 3). The integration of the results from these different analyses provided us a highly curated and trustworthy set of D. silvatica chemosensory-related transcripts.
Expression Profiling across Experimental Conditions
The pre-processed reads of each experimental condition (LEG#1, LEG#234, PALP, and REST) were back aligned to the final reference transcriptome using Bowtie version 1.0.0 (Langmead et al. 2009). We used RSEM 1.2.19 software to obtain read counts and TMM-normalized FPKMs (i.e., trimmed mean of M values-normalized fragments per kb of exon per million reads mapped) per transcript (Li and Dewey 2011). For the analysis, we consider that a gene is actually expressed when the FPKM values are >0.01, a reasonable cut-off given the low expression levels reported for other arthropod chemoreceptor proteins (Zhang et al. 2014). For the differential expression analysis, we considered that our data represent a single biological replicate (Robinson et al. 2010) and used EdgeR version 3.6.8 to calculate the negative binomial dispersion across conditions from the read counts of HK genes (Robinson et al. 2010). The P values from the differential expression analysis were adjusted for the false discovery rate (FDR; Benjamini and Hochberg 1995).
Phylogenetic Analyses
The quality of the MSA is critical to obtain a reliable phylogenetic reconstruction. This issue is very problematic in the face of highly divergent sequences, as in our case. To minimize this problem, we applied a profile-guided MSA approach based on highly curated Pfam core profiles, which generated MSAs with better TCS scores than other MSA approaches (Chang et al. 2014; J.-M. Chang et al. 2015). We used RAxML version 8.2.1 and the WAG protein substitution model with rate heterogeneity among sites to determine the phylogenetic relationships among the members of each chemosensory gene family in arthropods (Whelan and Goldman 2001; Stamatakis 2014). Node support was estimated from 500 bootstrap replicates. All phylogenetic tree images were created using the iTOL webserver (Letunic and Bork 2007). Trees were rooted according to available phylogenetic information; otherwise, we applied a midpoint rooting.
Results
Evaluation of the Best De Novo Assembly for D. s ilvatica Data
We obtained 441.8 million reads across the four experimental conditions, which dropped to 418.2 million (94.7%) after removing low-quality reads (table 1). We used the 98.4 million reads of the REST condition to evaluate the best de novo transcriptome assembler for our specific data. We found that among the assemblers using a single k-mer value of 25, SOAPdenovo-Trans and Trinity produced the largest number of contigs and the lowest N50 values (supplementary table S1, Supplementary Material online). The assembly based on Bridger provided the second best RSEM-EVAL score (after Trinity) but produced contigs with more positive BLAST hits against CEG and SwissProt proteins with a 100% alignment length filtering with an E-value of 10 − 3. Increasing the k-mer size had a disparate effect on the number of contigs and on the N50, but the resulting assemblies were generally worse than those generated using k-mer 25 (based on RSEM-EVAL scores and positive BLAST hits). Only the assemblies obtained in Bridger and Trinity with a k-mer of 31 outperformed their respective assemblies with a k-mer of 25. However, the multiple k-mer strategies implemented in Trans-Abyss and Oases yielded very different assembly qualities. Trans-Abyss produced a highly fragmented transcriptome (i.e., with a large number of very short contigs) that was clearly outperformed by Oases using the clustered option. Nevertheless, Oases performed worse than Bridger and Trinity (k-mer = 31) in terms of RSEM-EVAL scores and positive BLAST hits. Hence, although the Trinity assembly provided a lower RSEM-EVAL score, Bridger produced a very similar value of this parameter while performing better based on all other calculated statistics. Consequently, we selected Bridger with a k-mer of 31 as the best strategy for the de novo assembly of D. silvatica data and used the transcriptome from this software for further analyses.
Table 1.
Summary of RNA-Seq Data Assembly and Annotation
| PALP | LEG#1 | LEG#234 | REST | Total | Total aligned | |
|---|---|---|---|---|---|---|
| Total raw reads | 114,986,182 | 118,017,386 | 104,967,256 | 103,865,040 | 441,835,864 | 441,835,864 |
| GC (%) | 41.41 | 41.38 | 41.39 | 41.55 | 41.43 | 41.43 |
| Total qualified reads | 108,490,938 | 112,102,210 | 99,231,056 | 98,380,850 | 418,205,054 | 418,205,054 |
| Transcripts | 130,908 | 144,442 | 147,737 | 149,796 | 236,283 | 214,969 |
| Unigene transcripts (UT) | 93,283 | 104,004 | 106,966 | 109,335 | 170,846 | 154,427 |
| UT average length (in bp) | 1,027 | 956 | 943 | 932 | 702 | 751 |
| UT maximum length (in bp) | 26,709 | 26,709 | 26,709 | 26,709 | 26,709 | 26,709 |
| HK UT | 1,134 | 1,134 | 1,131 | 1,133 | 1,136 | 1,136 |
| CEG UT (CEG genes) | 766 (456) | 766 (457) | 775 (457) | 759 (457) | 807 (457) | 804 (457) |
| UT with GO annotation | 20,481 | 21,799 | 22,332 | 23,471 | 29,879 | 28,157 |
| UT with Interpro domain | 21,436 | 22,735 | 23,293 | 24,435 | 30,886 | 29,168 |
| UT with KEGG annotation | 3,313 | 3,409 | 3,444 | 3,599 | 3,895 | 3,817 |
| UT with functional annotation a | 21,567 | 22,874 | 23,438 | 24,600 | 31,091 | 29,359 |
| UT with genomic annotation b | 27,043 | 28,922 | 29,645 | 31,236 | 41,046 | 38,317 |
GO, Interpro or KEGG annotation.
GO, Interpro, KEGG annotation or BLAST hit.
The initial assembly from Bridger (using the reads from the four conditions) was formed by 236,283 contigs (after removing contaminant sequences), which decreased to 170,846 putative nonredundant transcripts after the clustering of isoforms (table 1). We identified 807 transcripts with significant BLAST hits against 457 out of the 458 CEGs, 454 of them with alignment lengths longer than the 60% of CEG target gene (234 with 100% of this length; supplementary table S2, Supplementary Material online). These results clearly demonstrate the completeness of the assembled transcriptome.
Functional Annotation of the D. s ilvatica Transcriptome
As expected, arthropodDB received the most significant positive BLAST hits with an E-value of 10 − 3 when using D. silvatica transcripts as queries (supplementary table S3, Supplementary Material online). Of these hits, 85% corresponded to chelicerate subjects; the spiders A. geniculata and S. mimosarum and the scorpion M. martensii were the most represented species (supplementary fig. S1, Supplementary Material online).
The most frequent GO terms associated with the D. silvatica transcripts were “metabolic” and “cellular processes” (biological process), as well as “binding” and “catalytic activities” (molecular function) (supplementary fig. S2, Supplementary Material online). Moreover, we found that 3,895 (out of the 29,879 transcripts with an associated GO term) showed significant positive BLAST hits against 136 different entries of the KEGG database (supplementary table S4, Supplementary Material online), with Purine metabolism (2,030 transcripts), Thiamine metabolism (1,053 transcripts) and Biosynthesis of antibiotics (454 transcripts including, e.g., some spider glutamate synthases and dehydrogenases) being the most represented pathways.
Condition-Specific Gene Expression Analysis
Our comparative analysis identified 57,282 transcripts expressed in all four conditions (37.1%) (fig. 2). The number of condition-specific transcripts in LEG#1, PALP and LEG#234 was rather similar (7,446, 6,000 and 8,605, respectively) and was much higher in REST (14,414), which is easily explained by the much larger number of tissues and physiological functions included in this condition. In the absence of separated biological replicates, we used the expression profile of HK genes to estimate the approximate dispersion of mean read counts across conditions to perform a rough differential expression analysis. The estimated dispersion across conditions of the 1,136 transcripts with significant positive BLAST hits to our set of HK genes (edgeR common dispersion value of 0.15) was used as the fold-change threshold for this analysis.
Fig. 2.—
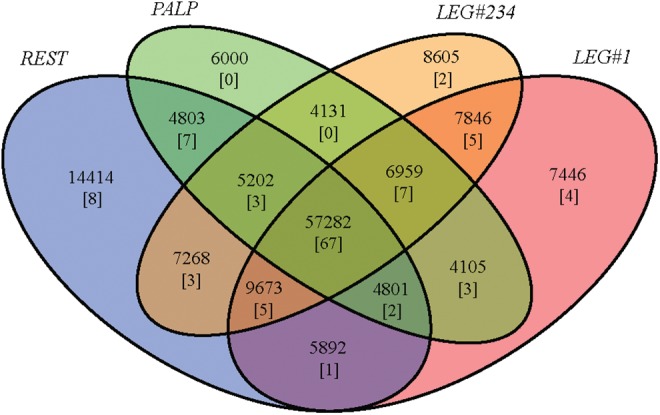
Venn diagram showing the total number of transcripts (154,427 transcripts) specifically expressed in each experimental condition and their intersections (red, orange, green and blue indicate LEG#1, LEG#234, PALP and REST, respectively). Numbers in brackets indicate putative chemosensory protein encoding transcripts (117 in total).
Our analyses show that LEG#1 and LEG#234 had rather similar transcriptomic profiles (supplementary fig. S3, Supplementary Material online). We found that only two transcripts were significantly overexpressed in LEG#1 and the other two in LEG#234; taking these two conditions together, there were 27 overexpressed transcripts, none annotated as a chemosensory gene. These results contrast with those obtained in PALP, where 174 transcripts were significantly overexpressed. However, again, none of these transcripts encoded an annotated chemosensory function; they were enriched in signal peptide encoding sequences (Fisher’s exact test, P value = 2.63×10 − 23), a feature characteristic of secreted proteins.
In addition, we found that the genes overexpressed in PALP were significantly enriched in the GO terms “metalloendopeptidase activity” (GO:0004222) and “proteolysis” (GO:0006508). In this specific tissue, these genes could be linked with the extra-oral digestion characteristic of these animals. However, we did not detect any GO term overrepresented in LEG or REST, and only 10 of the 27 genes significantly overexpressed in these structures had BLAST hits with an annotated sequence. Among these, we found genes encoding DNA-binding proteins, such as some transcription factors, hydrolases and proteins with transport activity.
Chemosensory Gene Families
To identify specific transcripts encoding chemosensory proteins in D. silvatica, we conducted additional exhaustive searches. We found many members of the Gr, Ir, Npc2 and Cd36-Snmp families, as well as putative distant homologs of insect OBPs and one uncharacterized protein family that may be involved in chemosensory function in this spider. Nevertheless, we failed to find homologs of the Csp gene family, which is present in the genome of other chelicerates. As expected, the D. silvatica transcriptome did not encode insect OR proteins nor their vertebrate functional counterparts (supplementary table S5 A, Supplementary Material online).
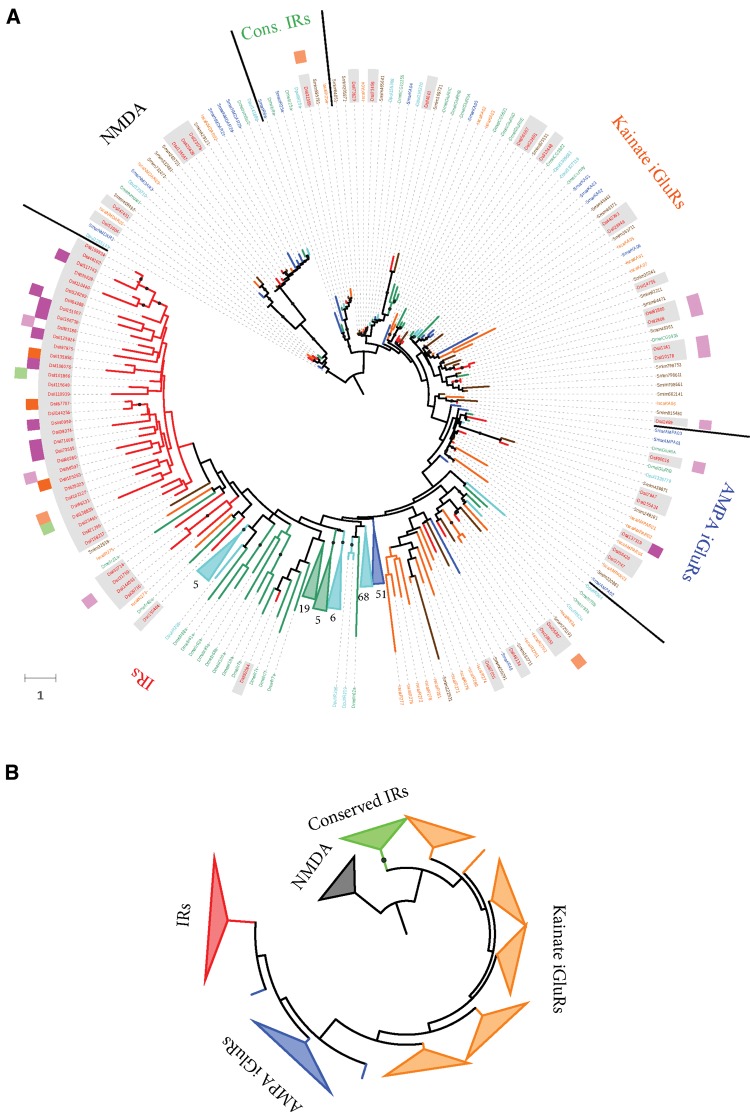
We identified 127 transcripts encoding IR/iGluR homologs (Ir transcripts), 57 exhibiting the specific domain signature of the ionotropic glutamate receptors (IPR001320). Some of these transcripts encoded some of the characteristic domains of the IR/iGluR proteins, such as the amino terminal (ATD-domain; PF01094), the ligand binding (LBD-domain; PF10613) and the ligand channel (LCD-domain; PF00060) (supplementary fig. S4, Supplementary Material online; see also Croset et al. 2010). Indeed, nine of them encoded all three domains, thus forming the typical complete iGluR structure, while 23 only had the two ligand-binding domains.
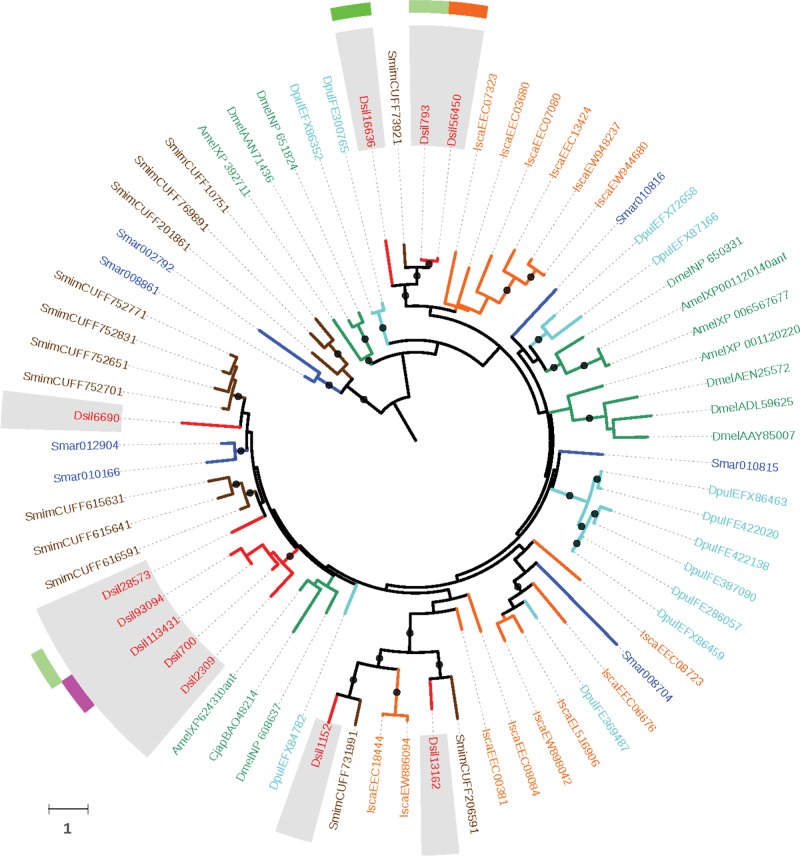
To understand the evolutionary diversification of the Ir/iGluR gene family in chelicerates, we carried out a protein domain-specific phylogenetic analysis. We used the information exclusively from the LCD domain because it is shared by all characterized arthropod IR/iGluR. For the analysis, we built an amino acid-based MSA including all D. silvatica transcript-coding LCD domains (70 transcripts) along with all reported sequences of this domain from D. melanogaster, D. pulex, S. maritima, I. scapularis, and S. mimosarum (i.e., in order to avoid large and unreadable trees, we included only one species per main arthropod lineage except for chelicerates, which were represented by a tick and a well annotated spider). We found that D. silvatica had representatives of all major IR/iGluR subfamilies, namely the AMPA, Kainate, NMDA (canonical iGluR subfamilies having all three Pfam domains), the two IR major subfamilies, the so called “conserved” IRs (encompassing the IR25a/IR8a members; having all three PFAM domains), and the remaining IR members (IR subfamily having only the LBD and LCD domains and that in Drosophila includes members with chemosensory function encompassing the so called “divergent” and the “antennal” IRs). In total, we identified 26 transcripts encoding canonical iGluR proteins plus another 44 encoding IRs (fig. 3 and supplementary fig. S5, Supplementary Material online), including a putative homolog of the highly conserved family of IR25a/IR8a proteins (transcript Dsil31989). Noticeably, this transcript is significantly overexpressed in LEG#1 with respect to REST (∼10 times more expression −logFC = 4; P < 0.01 after FDR), although it also shows 2 and 4 times more FPKM values with respect to PALP and LEG#234, respectively (supplementary table S5 B, Supplementary Material online).
Fig. 3.—
Maximum likelihood phylogenetic tree of the IR/iGluR proteins across arthropods. The tree is based on the MSA of the LCD domain (PF00060). (A) Sequences of Drosophila melanogaster, Daphnia pulex, Strigamia maritima, Ixodes scapularis, Stegodyphus mimosarum and Dysdera silvatica are depicted in green, light blue, dark blue, orange, brown and red, respectively. Additionally, the translation of the D. silvatica transcripts are shadowed in grey. Nodes with bootstrap support values >75% are shown as solid circles. Nodes with five or more sequences from the same species were collapsed; the actual number of collapsed branches is indicated in each case. The two surrounding circles provide information regarding the expression pattern of some D. silvatica genes. The most external circle indicates the genes specifically expressed in palps (PALP; in green), legs (both LEG#1 and LEG#234; in pink) and palps and legs (PALP, LEG#1 and LEG#234; in orange). The inner circle shows the genes overexpressed in these conditions using the same color codes but with two color intensities, one more intense color for overexpression levels >5× over REST and another lighter color for 2–5× overexpression values. The branch length scale is in numbers of amino acid substitutions per amino acid position. (B) Simplified phylogenetic tree highlighting the main Ir sub-families.
Our phylogenetic analysis uncovered a set of D. silvatica transcripts phylogenetically related to some D. melanogaster antennal IRs, such as the IR21a (Dsil32714), the IR40a (Dsil150464) and the IR93a (Dsil55987, Dsil29850 and Dsil48134) proteins. These transcripts, however, did not show any clear differential expression pattern in LEG#1 or PALP, while two of them were clearly overexpressed in REST. Moreover, similarly to what occurs in other arthropods, many nonconserved IRs formed a species-specific monophyletic clade (33 transcripts). Interestingly, 11 of these receptors were condition specific, and 8 were overexpressed (or showed at least 2 times more FPKMs) in the examined appendages (i.e., LEG#1, LEG#234 and PALP with respect to REST). Actually, LEG#1 was the expression condition with the highest number of different nonconserved Ir transcripts; only 14 of the 43 nonconserved Ir members were not expressed in this appendage (supplementary table S5 B, Supplementary Material online). Overall, the expression level of Irs (including conserved Irs) was lower than that of the iGluR transcripts.
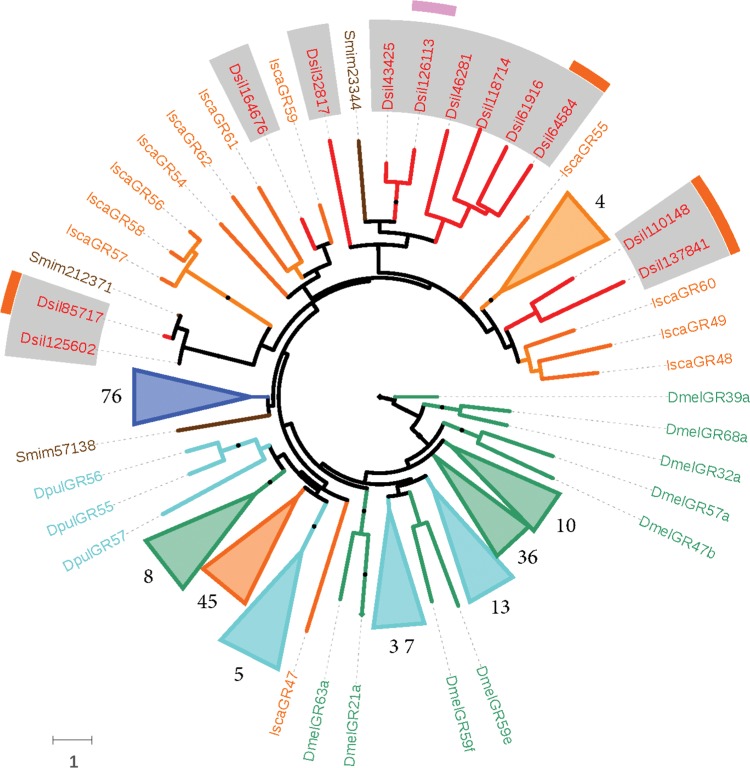
We further identified 12 transcripts encoding GR proteins (Gr transcripts), although only four of them had one of the two specific InterPro signatures that characterize this family (7m_7, IPR013604 and Trehalose receptor, IPR009318). In addition, these 12 Gr transcripts were phylogenetically related to members of this family characterized in the spider S. mimosarum and in the deer tick I. scapularis (fig. 4 and supplementary fig. S6, Supplementary Material online). The expression levels of D. silvatica Gr genes were considerably low compared both with the overall expression levels and with the expression levels of other chemosensory families (supplementary table S5 C, Supplementary Material online). Interestingly, only two Gr transcripts were condition specific (Dsil61916 and Dsil164676 in REST), and the other two were specifically expressed in both LEG#1 and PALP (Dsil110148 and Dsil137841). The remaining Gr transcripts showed variable gene expression profiles across conditions, with some genes having a wide expression pattern and others being more restricted to particular conditions (supplementary table S5 C, Supplementary Material online).
Fig. 4.—
Maximum likelihood phylogenetic tree of the GR proteins across arthropods. Species names, node support features and surrounding circles are colored as in figure 3.
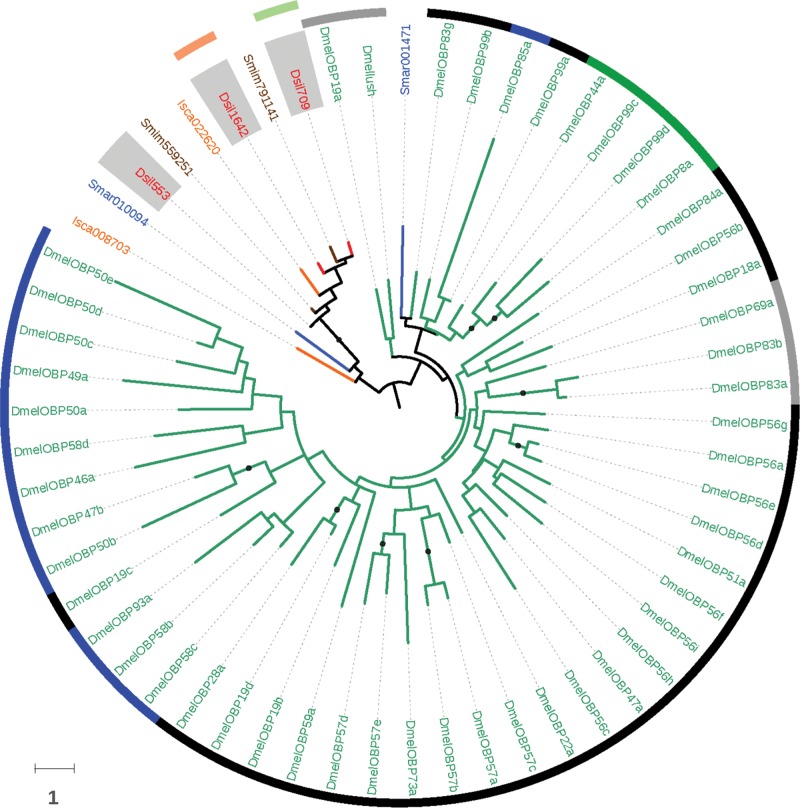
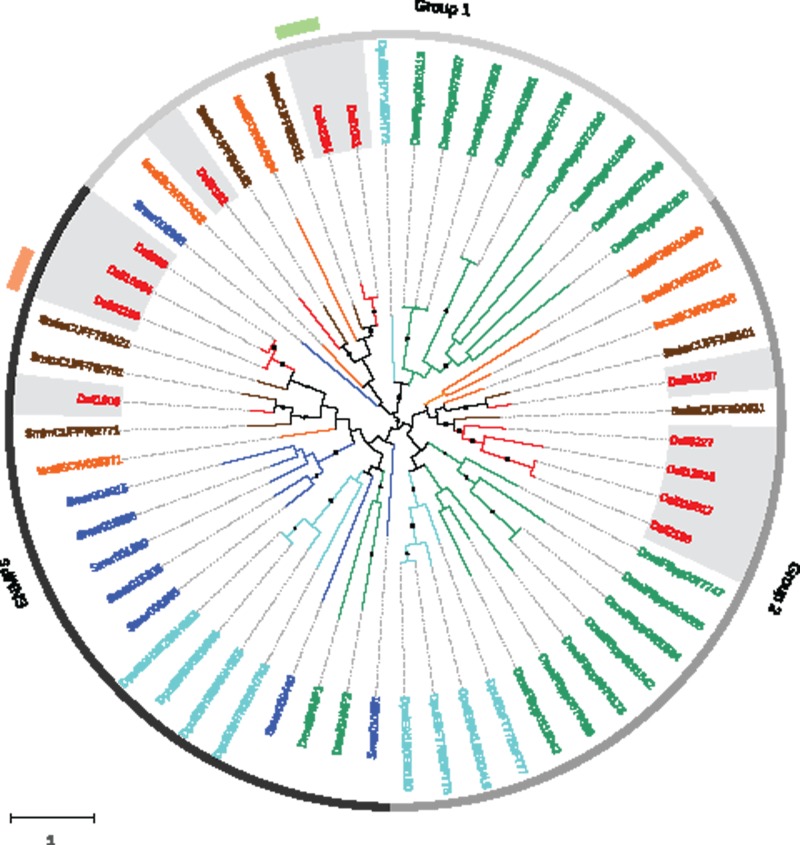
Our BLAST- and profile-based results revealed significant similarities between three spider transcripts and some insect members of the Obp family (with E-values between 10− 3 and 10− 5). The primary amino acid sequence and the cysteine pattern of the encoded proteins (hereafter designated OBP-like proteins) resembled those of OBPs and, one of them (Dsil553) showed a match to the PBP_GOBP InterPro domain (PBP_GOBP; IPR006170), uncovering a protein domain with folding features similar to those found in some insect OBPs. Using the three OBP-like sequences identified in the transcriptome of D. silvatica as a query in a BLASTp search against the NCBI-nr database (E-value of 10 − 3), we detected six additional members of this novel family in the genomes of S. mimosarum, I. scapularis and S. maritima (two copies in each genome; fig. 5) but none in the annotated proteomes of crustaceans. The MSA of the nine copies identified in noninsect species and all characterized members of the Obp family in D. melanogaster and Anopheles gambiae would suggest that the Obp-like family is distantly related to the Minus-C Obp subfamily. Despite the particularly low sequence similarity and the large differences in protein length (not only between OBP-like and insect OBPs but also among OBP members), three different MSAs built with different alignment algorithms, i.e., MAFFT with the option L-INS-I (Katoh and Standley 2013), PROMAL3D (Pei et al. 2008) and PSI-coffee (Chang et al. 2012), yielded exactly the same pattern of cysteine homology in the region of the GOBP-PBP domain. Accordingly, with these MSAs, OBP-like proteins lacked the same two structurally relevant cysteines as insect Minus-C OBPs (except the S. maritima protein Smar010094 in the MAFFT alignment; supplementary fig. S7, Supplementary Material online). These results, however, must be taken with caution due to the fact that some OBP-like as well as several insect OBPs show large amino or carboxy terminal domains outside the conserved OBP domain, some of them including extra cysteines. If these cysteines are not correctly aligned in their true homologous positions, the interpretation of the cysteine pattern of OBP-like proteins could be erroneous.
Fig. 5.—
Maximum likelihood phylogenetic relationships of spider OBP-like and insect OBP proteins. Species names, node support features and surrounding circles are colored as in figure 3. The inner circle labels the previously defined OBP phylogenetic subfamilies (Classic, Minus-C, Plus-C and ABPII in black, green, blue and grey, respectively).
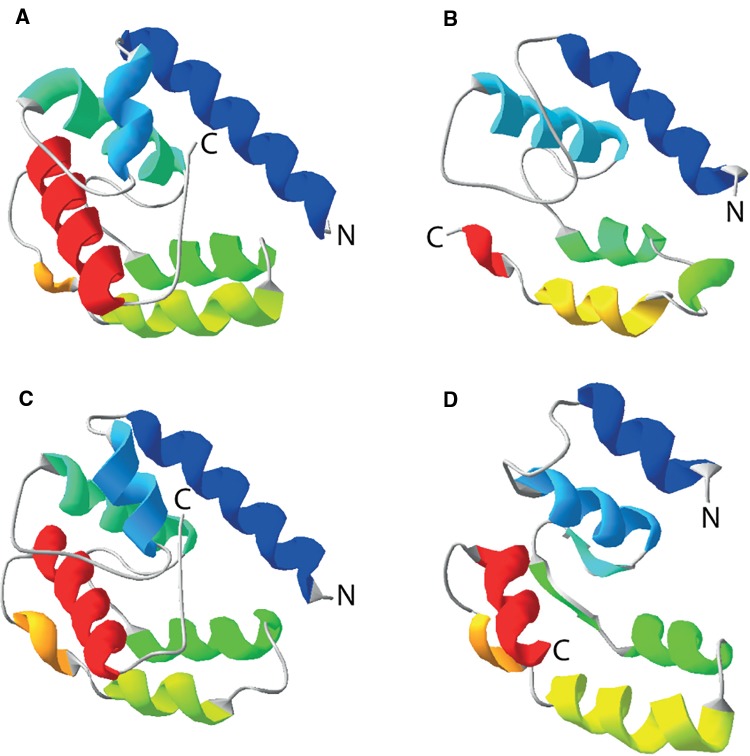
We built a 3D protein model of both the conceptual translation of one of the Obp-like transcripts identified in D. silvatica (Dsil553) and of the S. maritima protein Smar010094 using the Phyre2 web portal (Kelley et al. 2015). As expected, the predicted models showed a globular structure very similar to that found in insect OBPs (fig. 6). In fact, the top 10 structural templates identified by the software and, therefore, the one selected for the final modeling (A. gambiae proteins OBP20 and OBP4 for Dsil553 and Smar010094, respectively) were insect OBPs. In addition, the models showed a high confidence in the region corresponding to the GOBP-PBP domain (56% and 59% of the query sequences were modeled with 89.2% and 81.6% confidence by the single highest scoring template, respectively). Remarkably, the amino acid alignment between Smar010094 and OBP4, used as a guide by Pyre2 for building the 3D model of this S. maritima OBP-like protein, coincided with the PROMAL3D and Psi-Coffee alignments but not with the MAFFT one (see above). Hence, we hypothesize that, given the wide expression of spider OBP-like across the four experimental conditions (supplementary table S5 D, Supplementary Material online), these proteins, similar to those in insects, might be carriers of small soluble molecules acting in one or more physiological processes without ruling out a putative role in chemosensation.
Fig. 6.—
Predicted 3D structure of two OBP-like proteins. (A) Structure of Anopheles gambiae OBP20 (PDB 3V2L). (B) Structure of A. gambiae OBP4 (PDB 3Q8I). (C) 3D model of the protein encoded by the transcript Dsil553. (D) Predicted 3D model of the Strigamia maritima Smar010094 protein. PBD files were viewed and manipulated in Swiss-PdbViewer version 4.1 (Guex and Peitsch 1997).
We also identified 11 transcripts encoding putative NPC2 proteins, all of them having the characteristic IPR domain (MD-2-related lipid-recognition domain; IPR003172). The phylogenetic tree reconstructed from the MSA including these and other arthropod members of this family (including the members expressed in the antenna of A. mellifera and Camponotus japonicus (Ishida et al. 2014; Pelosi et al. 2014; fig. 7) uncovered a less dynamic gene family with neither large species-specific clades nor long branches. Nevertheless, internal node support was low and the precise phylogenetic relationships among arthropod NPC2s could not be determined with confidence. It is worth nothing, however, that this family underwent a moderate expansion in arthropods because it seems to be only one copy in both C. elegans and vertebrates. Only one putative D. silvatica Npc2 transcript was LEG#1 specific (Dsil113431), while two of them showed 11–4 times more FPKM in PALP (Dsil16636 and Dsil93094) and two others had 7 and 2 times more FPKM in LEG#1 and PALP than in REST (Dsil56450 and Dsil793), respectively (supplementary table S5 E, Supplementary Material online).
Fig. 7.—
Maximum likelihood phylogenetic tree of the NPC2 proteins across arthropods. Species names, node support features and surrounding circles are colored as in figure 3. Sequences from Apis mellifera and Camponotus japonicus are colored in green.
Finally, we identified 13 transcripts related to the Cd36-Snmp family, with 12 of them having the corresponding InterPro domain signature (CD36 antigen; IPR002159). Our phylogenetic analysis showed that D. silvatica had representatives of the three SNMP protein groups (Nichols and Vogt 2008; fig. 8), which would indicate that the origin of these subfamilies predated the diversification of the four major extant arthropod lineages. All four D. silvatica Snmp transcripts were similarly expressed in the four studied conditions, which would suggest either a nonchemosensory specific function of these proteins in spiders or a global general function within the chemosensory system (supplementary table S5 F, Supplementary Material online).
Fig. 8.—
Maximum likelihood phylogenetic tree of CD36-SNMP proteins across arthropods. Species names, node support features and surrounding circles are colored as in figure 3. The inner circle shows the different subfamilies.
A Novel Candidate Chemosensory Gene Family in Spiders
Furthermore, we conducted an exhaustive search on the 174 transcripts overexpressed in LEG#1 and PALP to try to identify putative novel, previously uncharacterized spider olfactory chemosensory families. For this, we first searched for gene families (groups of 4 or more similar sequences) by performing a clustering analysis of the 174 transcripts with CD-HIT (Fu et al. 2012); then, we searched for the presence of a signal peptide or for signs of trans-membrane helices in the identified families. We found one family (with five copies) in which one member had the molecular hallmark of a signal peptide; the absence of such a mark in the other four members could be due to the failure to detect full-length transcripts in these members (supplementary table S5 G, Supplementary Material online). Using these five sequences as queries in a BLAST search against the complete D. silvatica transcriptome, we further detected seven more members of this family. New BLAST searches using all 12 proteins as queries identified homologous copies in other spiders but not in the genomes of either other chelicerate lineages or nonchelicerate species. A preliminary phylogenetic analysis including all new identified sequences indicated that this family (supplementary fig. S8, Supplementary Material online) was highly dynamic, with several species-specific clades of CCPs (one of them including all D. silvatica copies) and no clear orthologous relationships across spiders. All these spider sequences, however, were annotated as uncharacterized proteins in these genomes. The MSA of the members of this novel family revealed a conserved cysteine pattern similar to that observed in insect OBPs and CSPs. However, unlike the OBP-like proteins, we could not obtain a reliable 3D protein model of a member of this family in the Phyre2 webserver. The server was unable to identify reasonable templates with large alignment coverage for the modeling (all templates with confidences >15 had an alignment coverage <7%). We then used I-TASSER suite (Yang et al. 2015) to try to find template proteins of similar folds as our D. silvatica queries. Although two of the identified threading templates were OBPs, some artificially designed proteins were also included in the modeling, generating five highly heterogeneous folding models, most of them with unacceptable C-scores. Nevertheless, some of the estimated folding models showed a compact global structure that, along with the presence of a signal peptide and the gene expression data, would suggest that the members this novel gene family could also acts as carriers of small soluble molecules, as insect OBP do (hereinafter we will refer to this novel family as the Ccp gene family for candidate carrier protein family).
Discussion
A High-Quality De Novo Assembly of the D. s ilvatica Transcriptome
The key step to obtain a high-quality transcriptome is selecting the best de novo assembly strategy and software. Nevertheless, because most assemblers have been developed for specific NGS platforms or tested using reduced data sets with limited taxonomic coverage, it is very difficult to predict their performance with disparate datasets (Martin and Wang 2011). Obtaining a high-quality transcriptome depends on factors such as the organism (which determines DNA complexity and heterozygosity levels), the read length and the sequencing depth. The best approach to determine the quality of different assemblies is to evaluate their accuracy (especially their completeness) in the context of a well-annotated, closely related reference genome (Marchant et al. 2015). Unfortunately, functionally annotated genomes of close relatives are usually not available for nonmodel organisms. In our case, the phylogenetically closest species with genome information, the spider L. reclusa, diverged from D. silvatica ∼200 Ma (Binford et al. 2008), which prevented any reliable evaluation. To circumvent this limitation, we used a combination of two strategies to evaluate the performance of five competing assemblers, one based on information of the transcriptome completeness (using CEG and SwissProt databases as subjects) and the other based on some statistics measuring the assembly quality (Li et al. 2014). Using this combined strategy and after evaluating 11 assembly scenarios, we were able to obtain a high-quality assembly that probably covers most of the D. silvatica transcriptome and that has a large proportion of full-length transcripts.
A Comprehensive Annotated Transcriptome That Uncovers a Surprising Gene Loss in Chelicerata
The functional annotation of a de novo assembled transcriptome from a nonmodel organism is a daunting task, being usually slow and computationally intensive. The large number of query sequences (transcripts) make similarity- and profile-based searches against general big databases, such as the NCBI-nr, very problematic, especially when using the free version of some software suites (e.g., BLAST2GO). Here, we used GHOSTZ instead of BLAST when searching against NCBI-nr, considerably reducing the computational time of the functional annotation step in >100 times, which is a relevant feature when testing assemblers in a comparative framework (i.e., a large number of independent annotations). Moreover, to increase the sensibility of the searches and reduce the computation time, we included only a representative set of phylogenetically close species to D. silvatica to build our specific databases (some annotated proteins are not yet available in NCBI-nr). Finally, we largely reduced the running time of the InterProScan searches (∼10 times) by using only the Pfam database (Finn et al. 2014) as a query without a substantial loss in the number of positive hits.
Despite the exhaustive annotation process, a high number of D. silvatica transcripts (81.8%) could not be functionally annotated. These percentages, however, are commonly obtained in RNA-Seq studies and can be attributable to different causes. First, nonannotated transcripts are significantly shorter than annotated ones (P value = 2.2×10 − 16), suggesting that many nonannotated transcripts are actually assembly errors or small fragments lacking any detectable protein domain signature (supplementary table S6, Supplementary Material online). Second, a fraction of these unannotated sequences could correspond to noncoding RNAs. Finally, the modest annotation of the genome of L. reclusa, the closest available relative to D. silvatica, could considerably reduce the success of our searches. In fact, an important number of D. silvatica transcripts without functional annotation (9,955 sequences) encoded proteins tagged as uncharacterized in the genome of L. reclusa.
A relevant result of our functional annotation of the D. silvatica transcriptome is the absence of a transcript encoding a Trehalase (KOG0602), the only gene of the CEG database not identified in the D. silvatica transcriptome. This gene seems to also be absent in the genomes of other chelicerates because we failed to detect it even using powerful profile-based approaches. Intriguingly, this protein is essential for insects (Shukla et al. 2015) not only because of its function as hydrolase but also for its involvement in the development of the optic lobe (Chen et al. 2014). Given that this gene is certainly present in the genome of all other major arthropod lineages as well as in the tardigrade H. dujardini and the nematode C. elegans, the most likely explanation for its absence is specific gene loss in the ancestor of chelicerates. The apparent absence of this gene in this lineage is interesting and clearly demands further investigation. The study of this gene loss, jointly with that of the set of uncharacterized proteins found in the D. silvatica transcriptome, will provide new insight into some important biological processes specific to chelicerates.
The Chemosensory Transcriptome of D. s ilvatica
Unlike our previous survey in the mygalomorph species M. calpeiana (Frías-López et al. 2015), here we identified several transcripts encoding members of chemosensory gene families in the four studied body parts, albeit with low expression levels. The different levels of success of the two studies could be related to the much higher sequencing depth (i.e., >10 Gbp sequenced per condition) of the D. silvatica RNA-Seq experiment.
As expected from the genome annotations of some chelicerate species, the transcriptome of D. silvatica did not contain genes related to the vertebrate chemoreceptors or odorant-binding protein families, ruling out the possibility that these or other similar families play any role in spider chemosensation. Similarly, we failed to detect members of the insect Or gene family, adding further evidence of the complete absence of this family in all arthropod lineages other than winged insects (Missbach et al. 2015). Moreover, despite the presence of members of the Csp gene family in some chelicerates and myriapods (Chipman et al. 2014; Qu et al. 2015; Gulia-Nuss et al. 2016), we did not identify any transcript encoding a protein with significant similarity to this family in D. silvatica. Although this negative result might be explained by sequencing or assembly limitations, Csp genes are also absent in all other spider genomes available in public repositories. We postulate that this gene family could have been lost early in the diversification of arachnids.
Candidate Spider Chemoreceptor Gene Families
Here, we identified a maximum of 12 transcripts encoding GR proteins (i.e., some of them may form part of the same gene), a number that may seem surprisingly small in comparison with the large number of Gr genes identified in the tick I. scapularis (62), the myriapod S. maritima (77) and the water flea D. pulex (58) genomes, for example. Nevertheless, given the underrepresentation of the chemosensitive hairs with respect to the total amount of tissue examined in each specific transcriptome, the identification and comprehensive annotation of the complete set of Gr genes are quite challenging in standard RNA-Seq studies (Zhang et al. 2014). In addition, some Gr genes do not necessarily have to be expressed at the precise moment (i.e., developmental stage or environmental condition) of the experiment (this can also be applied to all other chemosensory families). Therefore, the D. silvatica genome likely encodes many more members of this family, and the 12 transcripts found in this study are only a first preliminary subset of the gustatory repertoire of this spider. These molecules seem to be expressed across different spider body parts and some show specific expression in particular appendages, with groups of copies broadly expressed, other groups that are never found in particular appendages and others that show an opposite pattern of specificity. This combinatorial manner of expression is similar to that the described for the Grs in Drosophila, which would suggest analogous gustatory coding mechanisms in these two arthropods (Depetris-Chauvin et al. 2015; Joseph and Carlson 2015). The two phylogenetically related Gr genes specifically expressed in LEG#1 and PALP (Dsil110148 and Dsil137841) could be involved in the detection of some ecologically relevant signals, for example, partial pressure of CO2, in a similar way as some insect Gr specifically expressed in D. melanogaster antenna, although the proteins encoded by spider and insect transcripts are phylogenetically unrelated. In fact, all Gr transcripts detected in the D. silvatica transcriptome (including LEG#1 and PALP specific sequences) are members of a monophyletic group of chelicerate receptors for which we have no functional information. However, some Gr transcripts are also overexpressed or even exclusively expressed in the transcriptome of REST. The encoded proteins might participate in other, nonchemosensory physiological functions, as has also been observed in insects (Joseph and Carlson 2015). Even so, we cannot rule out that they actually act as chemoreceptors in other body structures, apart from palps and legs, such as in the mouthparts, which are included in REST transcriptome.
Unlike Grs, we have detected in D. silvatica a substantial number of sequences (127) encoding putative Ir transcripts, including a putative homolog of the conserved Ir subfamily Ir25a/Ir8a (Dsil31989). The phylogenetic analysis of the members of this family in arthropods clearly reflects the effect of the long-term birth-and-death process acting on most members of this family. Remarkably, this effect is almost unnoticeable in iGluR and in conserved IRs proteins, ratifying the marked differences in gene turnover rates between subfamilies. This highly dynamic evolution of nonconserved IR jointly with that reported for other proteins associated with contact chemoreception has been suggested as a proof of the high adaptive potential of the molecular components of the gustatory system in arthropods (see Torres-Oliva et al. 2016, and references therein). Interestingly, some of the 10 nonconserved IRs not included in the D. silvatica-specific clade are phylogenetically related to some D. melanogaster antennal IRs, including one member that presumably plays an important role in thermosensation (IR21a). Nevertheless, the expression profiles of these five transcripts do not provide clues regarding their possible role in spider chemosensation (i.e., they do not show any specific gene expression pattern across conditions). Although the putative spider homolog of the Ir25a/Ir8a subfamily is also expressed in all four conditions, it is much more abundant in PALP, LEG#1 and LEG#234, and even significantly overexpressed in LEG#1 with respect to REST. The IR25a and IR28a proteins are widely expressed in Drosophila olfactory sensilla (and in olfactory organs of other arthropods; Croset et al. 2010) and have been involved in the trafficking to the membrane of the other IR and in a co-receptor function of food-derived chemicals and humidity and temperature preferences. Thus, our results indicate that the first pair of legs of spiders could be relevant for the detection of amines and/or aldehydes as well as for determining favorable ranges of certain environmental variables (Silbering et al. 2011; Min et al. 2013; Enjin et al. 2016). Finally, and similar to that observed in for Gr transcripts, some members of the nonconserved Ir subfamily are also detected in REST, further supporting their involvement in other nonchemosensory functions or, alternatively, the presence of chemosensory structures in body parts other than legs or palps.
Evolution of the IR Family in Arthropods
Since our phylogenetic analysis includes highly diverged sequences, we applied for first time domain-specific HMM profiles to guide the MSA of chemosensory families. This strategy has been especially useful for the Ir/iGluR families, exploiting the evolutionary information of the conserved ligand channel domain (LCD domain) clearly shared by all known members. The inferred tree mirrors the same focal phylogenetic groups obtained in previous works (Croset et al. 2010). Most tree reconstructions show that (1) the Kainate and AMPA proteins are closely related, and AMPA likely a derived linage, (2) the subfamily of the conserved IRs is the sister group of these Kainate/AMPA receptors, and (3) NMDA sequences represent the first offshoot. However, there are some important differences between the present study and findings regarding the putative origin of the nonconserved IRs. This group of IRs, which forms a supported monophyletic group in all tree reconstructions, is more closely related to non-NMDA receptors than to the remaining iGluRs in our tree, which could indicate that they originated from a Kainate- or AMPA-like receptor. Nevertheless, the poor support of some internal nodes, probably due to alignment artifacts caused by the diverse domain structure of Ir/iGluR families, precludes making definitive conclusions about the origin of these highly divergent receptors.
Novel Classes of Candidate Transport Proteins in Chelicerates
Pelosi et al. (Pelosi et al. 2014) proposed that some members of the Npc2 family might be involved in the transport and solubilization of semiochemicals in noninsect arthropods, constituting an alternative to the insect OBP and CSP proteins involved in the peripheral events of olfaction. Here, we show that the spider D. silvatica has a similar repertoire of Npc2 genes to that found in other surveyed arthropods, which seems to be expanded in arachnids. We identified one member of this family specifically expressed in LEG#1 that may be a good candidate to participate in odor detection in spiders; this transcript, however, showed a relatively low expression level, in contrast to the very high expression levels observed in insect Obp and Csp genes. Although the remaining members of the Npc2 family might also have other chemoreceptor functions in Dysdera, most of them probably perform other important physiological functions, such as cholesterol lipid binding and transport, which is the known function of these proteins in vertebrates (Storch and Xu 2009).
One unexpected and remarkable result is the expression in D. silvatica of at least three genes encoding proteins with a secondary structure, conserved cysteine pattern (revealed in the MSAs that include insect OBPs and characteristic of the Minus-C subfamily) and predicted folding similar to that of insect OBPs. In fact, our searches using these newly identified OBP-like proteins as a query revealed that chelicerates and myriapods, but not crustacean or insects, have some copies of this family. In the absence of confirmation by functional experiments and structural data, these results suggest that the Obp superfamily was already present in the arthropod ancestor. We cannot confirm whether putative ancestors were actually members of the Minus-C subfamily because this group of proteins is polyphyletic in the OBP tree (Vieira and Rozas 2011). Nevertheless, the fact that chelicerate and myriapod genomes only carry Minus-C Obp genes supports them as the ancestral arthropod Obp. In D. melanogaster, the Minus-C Obps are highly expressed in several tissues other than the head, including adult carcass, testis, male accessory glands, spermatheca and some larval tissues (data from FlyAtlas project; Chintapalli et al. 2007). The wide expression levels of OBP-like genes across all four experimental conditions, together with their low gene turnover rates in chelicerates, also indicate essential and multiple functional roles of these putative small soluble carriers, regardless of their possible function in the chemosensory system.
Lastly, the newly identified Ccp family encodes a protein with a clear signal peptide that shows similar folding characteristics to those of insect OBPs. Interestingly, half of their members are overexpressed in the proposed spider olfactory organs. In this case, however, we only detected homologous copies in the genomes of arachnids, where the products are annotated as uncharacterized proteins. Thus, both the NPC2 copy and the proteins encoded by the Ccp family are good candidate chelicerate counterparts of the insect OBP and the CSP proteins, and their specific function clearly deserves further exploration.
In this study, we report the first comprehensive comparative transcriptomic analysis across different body structures of a spider, including those that most likely carry the chemosensory hairs. Our results indicate that, as in other noninsect arthropods, gustatory and ionotropic receptor families are the best candidate peripheral chemoreceptors in chelicerates. Additionally, we found some noteworthy differences in the specific pattern of gene expression of the members of these chemosensory families across different body structures, some of them involving the putative olfactory system-containing organs, which can indicate some specialization of chemosensory structures across the body of D. silvatica. In addition, we identified a protein family in chelicerates that seems to be distantly related to the insect Obp family and have characterized a new gene family of small secreted soluble proteins analogous to the insect OBPs or CSPs that could act as molecular carriers in this species. Finally, we provide the first complete and functionally annotated transcriptome of a polyphagous predator species of the genus Dysdera, which will provide valuable information for further studies on this group, and a list of candidate genes suitable for further functional dissection. Our results will help better establish the specific role and sensory modality of each of these new identified genes and gene families in spiders while providing new insight into the origin and evolution of the molecular components of the chemosensory system in arthropods.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the Ministerio de Economía y Competitividad of Spain (BFU2010-15484, CGL2012-36863 and CGL2013-45211) and the Comissió Interdepartamental de Recerca I Innovació Tecnològica of Catalonia, Spain (2009SGR-1287, 2014SGR-1055 and 2014SGR1604). J.V. was supported by an FPI grant (Ministerio de Economía y Competitividad of Spain, BES-2014-068437), C.F.-L. by an IRBio fellowship, and A.S.-G. by a Beatriu de Pinós grant (Generalitat de Catalunya, 2010-BP-B 00175), and J.R. and M.A.A. were partially supported by ICREA Academia (Generalitat de Catalunya). We acknowledge the Garajonay National Parks for granting collection permits and helping with lodging and logistics during fieldwork.
Literature Cited
- Adams MD, et al. 2000. The genome sequence of Drosophila melanogaster . Science 287:2185–2195. [DOI] [PubMed] [Google Scholar]
- Altschul S. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Ashburner M, et al. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 57:289–300. [Google Scholar]
- Binford GJ, et al. 2008. Phylogenetic relationships of Loxosceles and Sicarius spiders are consistent with Western Gondwanan vicariance. Mol Phylogenet Evol. 49:538–553. [DOI] [PubMed] [Google Scholar]
- Birol I, et al. 2009. De novo transcriptome assembly with ABySS. Bioinformatics 25:2872–2877. [DOI] [PubMed] [Google Scholar]
- Cao Z, et al. 2013. The genome of Mesobuthus martensii reveals a unique adaptation model of arthropods. Nat Commun. 4:2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveira AM, Jackson RR. 2012. Love is in the air: olfaction-based mate-odour identification by jumping spiders from the genus Cyrba . J Ethol. 31:29–34. [Google Scholar]
- Chang J-M, Di Tommaso P, Lefort V, Gascuel O, Notredame C. 2015. TCS: a web server for multiple sequence alignment evaluation and phylogenetic reconstruction. Nucleic Acids Res. 43:W3–W6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J-M, Di Tommaso P, Notredame C. 2014. TCS: a new multiple sequence alignment reliability measure to estimate alignment accuracy and improve phylogenetic tree reconstruction. Mol Biol Evol. 31:1625–1637. [DOI] [PubMed] [Google Scholar]
- Chang JM, Di Tommaso P, Taly JF, Notredame C. 2012. Accurate multiple sequence alignment of transmembrane proteins with PSI-Coffee. BMC Bioinformatics 13(Suppl 4):S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Z, et al. 2015. Bridger: a new framework for de novo transcriptome assembly using RNA-seq data. Genome Biol. 16:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EA, et al. 2014. Effect of RNA integrity on uniquely mapped reads in RNA-Seq. BMC Res Notes 7:753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JAT. 2007. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 39:715–720. [DOI] [PubMed] [Google Scholar]
- Chipman AD, et al. 2014. The first myriapod genome sequence reveals conservative arthropod gene content and genome organisation in the centipede Strigamia maritima . PLoS Biol. 12:e1002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TH, et al. 2014. Multi-tissue transcriptomics of the black widow spider reveals expansions, co-options, and functional processes of the silk gland gene toolkit. BMC Genomics 15:365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbourne JK, et al. 2011. The ecoresponsive genome of Daphnia pulex . Science 331:555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, et al. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. [DOI] [PubMed] [Google Scholar]
- Corey EA, Bobkov Y, Ukhanov K, Ache BW. 2013. Ionotropic crustacean olfactory receptors. PLoS One 8:e60551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croset V, et al. 2010. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 6:e1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depetris-Chauvin A, Galagovsky D, Grosjean Y. 2015. Chemicals and chemoreceptors: ecologically relevant signals driving behavior in Drosophila . Front Ecol Evol. 3:41. [Google Scholar]
- Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput. Biol. 7:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjin A, et al. 2016. Humidity sensing in Drosophila . Curr Biol. 26:1352–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, et al. 2014. Pfam: the protein families database. Nucleic Acids Res. 42:D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foelix RF, Chu-Wang IW. 1973. The morphology of spider sensilla. II. Chemoreceptors. Tissue Cell 5:461–478. [DOI] [PubMed] [Google Scholar]
- Foelix RF, Rast B, Peattie AM. 2012. Silk secretion from tarantula feet revisited: alleged spigots are probably chemoreceptors. J Exp Biol. 215:1084–1089. [DOI] [PubMed] [Google Scholar]
- Foelix RF. 1970. Chemosensitive hairs in spiders. J Morphol. 132:313–333. [DOI] [PubMed] [Google Scholar]
- Frías-López C, et al. 2015. Comparative analysis of tissue-specific transcriptomes in the funnel-web spider Macrothele calpeiana (Araneae, Hexathelidae). Peer J. 3:e1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbić M, et al. 2011. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 479:487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh-Lunow KC, Getahun MN, Grosse-Wilde E, Hansson BS. 2014. Expression of ionotropic receptors in terrestrial hermit crab’s olfactory sensory neurons. Front Cell Neurosci. 8:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723. [DOI] [PubMed] [Google Scholar]
- Gulia-Nuss M, et al. 2016. Genomic insights into the Ixodes scapularis tick vector of Lyme disease. Nat Commun. 7:10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, et al. 2014. De novo transcript sequence reconstruction from RNA-Seq: reference generation and analysis with Trinity. Nat Protoc. 8:1–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Marin J, Suleski M, Paymer M, Kumar S. 2015. Tree of life reveals clock-like speciation and diversification. Mol Biol Evol. 32:835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida Y, et al. 2014. Niemann-Pick type C2 protein mediating chemical communication in the worker ant. Proc Natl Acad Sci U S A. 111:3847–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph RM, Carlson JR. 2015. Drosophila chemoreceptors: a molecular interface between the chemical world and the brain. Trends Genet. 31:683–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. 2000. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaupp UB. 2010. Olfactory signalling in vertebrates and insects: differences and commonalities. Nat Rev Neurosci. 11:188. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkness EF, et al. 2010. Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc Natl Acad Sci U S A. 107:12168–12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305:567–580. [DOI] [PubMed] [Google Scholar]
- Kronestedt T. 1979. Study on chemosensitive hairs in wolf spiders (Araneae, Lycosidae) by scanning electron microscopy. Zool Scr. 8:279–285. [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le H-S, Schulz MH, McCauley BM, Hinman VF, Bar-Joseph Z. 2013. Probabilistic error correction for RNA sequencing. Nucleic Acids Res. 41:e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128. [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, et al. 2014. Evaluation of de novo transcriptome assemblies from RNA-Seq data. Genome Biol. 15:553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant A, et al. 2015. Comparing de novo and reference-based transcriptome assembly strategies by applying them to the blood-sucking bug Rhodnius prolixus . Insect Biochem Mol Biol. 69:25–33. [DOI] [PubMed] [Google Scholar]
- Martin J. a, Wang Z. 2011. Next-generation transcriptome assembly. Nat Rev Genet. 12:671–682. [DOI] [PubMed] [Google Scholar]
- Min S, Ai M, Shin SA, Suh GSB. 2013. Dedicated olfactory neurons mediating attraction behavior to ammonia and amines in Drosophila. Proc Natl Acad Sci U S A. 110:E1321–E1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missbach C, Vogel H, Hansson BS, Groβe-Wilde E. 2015. Identification of odorant binding proteins and chemosensory proteins in antennal transcriptomes of the jumping bristletail Lepismachilis y-signata and the firebrat Thermobia domestica: evidence for an independent OBP-OR origin. Chem Senses 40:615–626. [DOI] [PubMed] [Google Scholar]
- Mita K, et al. 2004. The genome sequence of silkworm, Bombyx mori . DNA Res. 11:27–35. [DOI] [PubMed] [Google Scholar]
- Mitchell A, et al. 2014. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 43: D213–D221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagné N, de Fouchier A, Newcomb RD, Jacquin-Joly E. 2015. Advances in the identification and characterization of olfactory receptors in insects. Prog Mol Biol Transl Sci. 130:55–80. [DOI] [PubMed] [Google Scholar]
- Nelson XJ, Warui CM, Jackson RR. 2012. Widespread reliance on olfactory sex and species identification by lyssomanine and spartaeine jumping spiders. Biol J Linn Soc. 107:664–677. [Google Scholar]
- Nichols Z, Vogt RG. 2008. The SNMP/CD36 gene family in Diptera, Hymenoptera and Coleoptera: Drosophila melanogaster, D. pseudoobscura, Anopheles gambiae, Aedes aegypti, Apis mellifera, and Tribolium castaneum . Insect Biochem Mol Biol. 38:398–415. [DOI] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Korf I. 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23:1061–1067. [DOI] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Ning Z, Keane T, Korf I. 2009. Assessing the gene space in draft genomes. Nucleic Acids Res. 37:289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7:e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Kim BH, Grishin VN. 2008. PROMALS3D: a tool for multiple sequence and structure alignment. Nucleic Acids Res. 36:2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi P, Iovinella I, Felicioli A, Dani FR. 2014. Soluble proteins of chemical communication: an overview across arthropods. Front Physiol. 5:320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi P, Zhou J-J, Ban LP, Calvello M. 2006. Soluble proteins in insect chemical communication. Cell Mol Life Sci. 63:1658–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi P. 1996. Perireceptor events in olfaction. J Neurobiol. 30:3–19. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. [DOI] [PubMed] [Google Scholar]
- Posnien N, et al. 2014. A comprehensive reference transcriptome resource for the common house spider Parasteatoda tepidariorum . PLoS One 9:e104885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu S-X, Ma L, Li H-P, Song J-D, Hong X-Y. 2015. Chemosensory proteins involved in host recognition in the stored food mite Tyrophagus putrescentiae . Pest Manag Sci. 72(8):1508–1516. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota-Stabelli O, Daley AC, Pisani D. 2013. Molecular timetrees reveal a Cambrian colonization of land and a new scenario for ecdysozoan evolution. Curr Biol. 23:392–398. [DOI] [PubMed] [Google Scholar]
- Sánchez-Gracia A, Vieira FG, Almeida FC, Rozas J. 2011. Comparative genomics of the major chemosensory gene families in arthropods. Encycl Life Sci. 3:476–490. [Google Scholar]
- Sanggaard KW, et al. 2014. Spider genomes provide insight into composition and evolution of venom and silk. Nat Commun. 5:3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz MH, Zerbino DR, Vingron M, Birney E. 2012. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28:1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag SR, et al. 2001. Expression mosaic of odorant-binding proteins in Drosophila olfactory organs. Microsc Res Tech. 55:297–306. [DOI] [PubMed] [Google Scholar]
- Shukla E, Thorat LJ, Nath BB, Gaikwad SM. 2015. Insect trehalase: physiological significance and potential applications. Glycobiology 25:357–367. [DOI] [PubMed] [Google Scholar]
- Silbering AF, et al. 2011. Complementary function and integrated wiring of the evolutionarily distinct Drosophila olfactory subsystems. J Neurosci. 31:13357–13375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch J, Xu Z. 2009. Niemann-Pick C2 (NPC2) and intracellular cholesterol trafficking. Biochim Biophys Acta. 1791:671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Kakuta M, Ishida T, Akiyama Y. 2014. Faster sequence homology searches by clustering subsequences. Bioinformatics 31:1183–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Oliva M, Almeida FC, Sánchez-Gracia A, Rozas J. 2016. Comparative genomics uncovers unique gene turnover and evolutionary rates in a gene family involved in the detection of insect cuticular pheromones. Genome Biol Evol. 8:1734–1747. [Google Scholar]
- Vieira FG, Rozas J. 2011. Comparative genomics of the odorant-binding and chemosensory protein gene families across the arthropoda: origin and evolutionary history of the chemosensory system. Genome Biol Evol. 3:476–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt RG, Riddiford LM. 1981. Pheromone binding and inactivation by moth antennae. Nature 293:161–163. [DOI] [PubMed] [Google Scholar]
- Whelan S, Goldman N. 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 18:691–699. [DOI] [PubMed] [Google Scholar]
- Whiteman NK, Pierce NE. 2008. Delicious poison: genetics of Drosophila host plant preference. Trends Ecol Evol. 23:473–478. [DOI] [PubMed] [Google Scholar]
- World Spider Catalog. 2016. World Spider Catalog. Nat. Hist. Museum Bern:online at http://wsc.nmbe.ch; version 17.0.
- Xie Y, et al. 2014. SOAPdenovo-Trans: de novo transcriptome assembly with short RNA-Seq reads. Bioinformatics 30:1660–1666. [DOI] [PubMed] [Google Scholar]
- Yang J, et al. 2015. The I-TASSER suite: protein structure and function prediction. Nat Methods 12:7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zheng Y, Li D, Fan Y. 2014. Transcriptomics and identification of the chemoreceptor superfamily of the pupal parasitoid of the oriental fruit fly, Spalangia endius Walker (Hymenoptera: Pteromalidae). PLoS One 9:e87800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.