Abstract
The western honey bee, Apis mellifera, provides critical pollination services to agricultural crops worldwide. However, despite substantial interest and prior investigation, the early evolution and subsequent diversification of this important pollinator remain uncertain. The primary hypotheses place the origin of A. mellifera in either Asia or Africa, with subsequent radiations proceeding from one of these regions. Here, we use two publicly available whole-genome data sets plus newly sequenced genomes and apply multiple population genetic analysis methods to investigate the patterns of ancestry and admixture in native honey bee populations from Europe, Africa, and the Middle East. The combination of these data sets is critical to the analyses, as each contributes samples from geographic locations lacking in the other, thereby producing the most complete set of honey bee populations available to date. We find evidence supporting an origin of A. mellifera in the Middle East or North Eastern Africa, with the A and Y lineages representing the earliest branching lineages. This finding has similarities with multiple contradictory hypotheses and represents a disentangling of genetic relationships, geographic proximity, and secondary contact to produce a more accurate picture of the origins of A. mellifera. We also investigate how previous studies came to their various conclusions based on incomplete sampling of populations, and illustrate the importance of complete sampling in understanding evolutionary processes. These results provide fundamental knowledge about genetic diversity within Old World honey bee populations and offer insight into the complex history of an important pollinator.
Keywords: genomics, population genetics, ancestry, Apis mellifera
Introduction
The western honey bee, Apis mellifera, is the most important insect pollinator of agricultural crops worldwide. Numerous food commodities (e.g. almond, apple, watermelon) rely heavily or exclusively on honey bees for fruit, vegetable, or seed production (Klein et al. 2007). In fact, the dependence of agricultural activities on honey bee pollination services has increased during the last few decades (Aizen et al. 2009). In the United States alone, the value of honey bee pollination services is estimated between 10 and 14 billion dollars annually (Calderone 2012). In addition, a variety of honey bee-derived products (e.g. honey, wax, pollen) trade as international commodities (vanEngelsdorp and Meixner 2010). However, despite the critical importance of honey bee genetic diversity for breeding practices and food security, our current understanding of the demographic history and evolutionary origin of contemporary (native and managed) populations of honey bees remains unclear.
The genus Apis contains ten distinct species, most of which are distributed across Asia (Arias and Sheppard 2005). The western honey bee, A . mellifera, was historically distributed throughout sub-Saharan Africa, Europe, parts of western Asia, and the Middle East, and is currently the only species of honey bee that has undergone substantial domestication. This species is thought to have split from its close relative, A. cerana, between 6 and 25 million years ago, when it expanded westward to colonize parts of Asia, Europe, and Africa (Sheppard and Meixner 2003; Ramirez et al. 2010). Subsequently, as European settlers colonized different parts of the globe, different lineages of A. mellifera were transported and established along with elaborate beekeeping practices, resulting in the naturalization of multiple interbreeding lineages (Sheppard 1989).
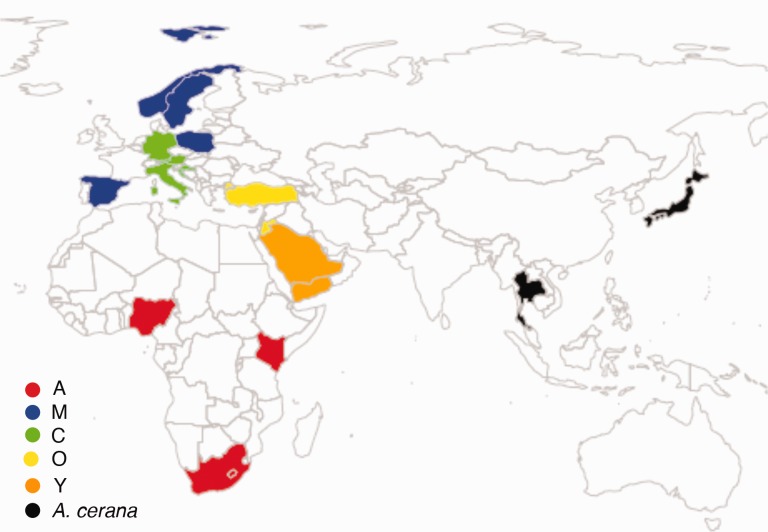
Across its native range, A. mellifera exhibits substantial genetic and phenotypic variation of both behavioral and morphological traits (Ruttner 1988). At least 26 morphologically and geographically distinct subspecies have been identified, and geometric morphometric analyses coupled with genetic studies have strongly supported the existence of four distinct lineages (hereafter called M, C, O, and A) (fig. 1) (Ruttner 1988; Franck et al. 2000; Whitfield et al. 2006). More recently a fifth lineage (Y) was identified from northeastern Africa and the Middle East (Franck et al. 2001), and a possible additional (sixth) lineage was reported from Syria (Alburaki et al. 2011; Alburaki et al. 2013). However, the relationships among these lineages, and the evolutionary trajectories that gave rise to their diversification into geographically distinct populations, remain unclear. In particular, the geographic region consisting of the Middle East and Northeastern Africa contains several contact zones between the A, O, Y, and potentially, other lineages.
Fig. 1.—
Collection locations for populations used in this study. Map generated with rworldmap in R (South 2011).
Multiple scenarios have been put forward to explain the demographic history and evolutionary origin of extant A. mellifera lineages. The debate arises, in part, due to conflicting pieces of evidence that variously support the origin of A. mellifera in Asia, the Middle East, or Africa (Han et al. 2012). Using nuclear and mitochondrial markers, Arias and Sheppard (2005) indicated that A. mellifera has low sequence divergence, which supports a scenario of a relatively recent diversification. In addition, A. mellifera is substantially diverged from its sister taxa, all of which are native to Asia. Further contradictory evidence arises from the observation that African populations exhibit comparatively higher levels of genetic diversity (total number of SNPs) as well as higher mean nucleotide diversity values (π) (Han et al. 2012;Wallberg et al. 2014) relative to all other A. mellifera populations. Together, these observations have been used to suggest Africa as the center of origin of genetic lineages of A. mellifera. However, additional confusion has arisen due to the geographic proximity of several A. mellifera populations that are genetically separated from each other such as the C and M lineages in Europe and the O and Y lineages in the Middle East (Franck et al. 2001). A recent microsatellite analysis of A. mellifera collected in Syria, Lebanon, and Iraq further complicate the picture with unclear placement of these individuals between the O and A lineages (Alburaki et al. 2013).
In the last decade, several studies have made attempts to resolve this issue using population genomic tools. Whitfield et al. ( 2006) generated 1136 SNPs from the nuclear genome and identified Africa as the origin of A. mellifera, with the M, C, and O lineages representing between two and three expansions from ancestral African populations. However, reanalysis of this data set by Han et al. (2012) did not find strong support for any particular hypothesis, but instead, suggested an origin close to the region where Asian sister species are found. A more recent analysis by Wallberg et al. (2014) also found no evidence for an African origin of A. mellifera and concurred with the Han et al. 2012 study as more consistent with their findings. However, none of these studies included samples from populations spanning the entire range of lineages in A. mellifera. In particular, samples that are geographically intermediate to A. mellifera and its sister species (A. cerana) are clearly necessary to correctly infer the demographic and evolutionary history of this species (Sheppard and Meixner 2003; Chen et al. 2016).
Here, we combine two publicly available data sets (Harpur et al. 2014; Wallberg et al. 2014) with some additional newly sequenced individuals to produce the most comprehensive whole genome data set to date for A. mellifera. The combined data set includes individuals from five of the major A. mellifera lineages (A, C, M, O, and Y) and spans a large geographic range, including Africa, Europe, and the Middle East (fig. 1). Therefore, this data set is uniquely able to address the evolutionary origins of A. mellifera and clarifies some of the earlier confusion about the relationships between the major A. mellifera lineages. Our analysis does not identify the precise origin of A. mellifera, but it does resolve some of the apparent contradictions in the literature with respect to the relationships between the major lineages and suggests that an origin in the Middle East or Northeastern Africa is most likely. In addition, our analysis attempts to distinguish between the geographic hypotheses proposed for the origin of A. mellifera and the hypotheses for the intraspecific relationships among the major lineages within A. mellifera. While the geographic location of the origin of A. mellifera that we hypothesize is similar to that proposed by Ruttner et al. (1978) based on morphological analysis, our hypothesis is more similar to that advanced by Whitfield et al. (2006) with respect to the relationships between the A, C, M, and O A. mellifera lineages. We believe that we have disentangled a particularly convoluted evolutionary history and, in addition to our biological conclusions, we also discuss how subsets of the data can produce substantively different conclusions.
We additionally harness this extensive data set to identify patterns of gene differentiation between populations to identify potential patterns of local adaptation. We detected strong evidence of admixture between populations of A. mellifera as well as evidence of gene ontology categories, such as sensory transduction and transmembrane helix, which show evidence of repeated differentiation between lineages. We also find a significant increase in SNPs in exonic regions of the genome, including dozens of non-synonymous SNPs that show differentiation between lineages in some important and well-studied A. mellifera genes, such as vitellogenin and the major royal jelly proteins. This analysis provides a better understanding of the genetic diversity and evolutionary history of honey bee lineages, and the biological conclusions drawn here can be applied to honey bee breeding practices as well as honey bee health.
Materials and Methods
Data Sets
We downloaded whole genome fastq files for two publicly available A. mellifera data sets. The first data set was described in Harpur et al. (2014) and is available from the Sequence Read Archive (PRJNA216922). This data set consists of 39 A. mellifera samples and one A. cerana outgroup sample sequenced in an Illumina HiSeq 2000. Ten of these A. mellifera samples belong to the Y group, nine belong to the C group, nine belong to the M group, and 11 belong to the A group. The one A. cerana individual was from Thailand. The second data set was described in Wallberg et al. (2014) and is available from the Sequence Read Archive (PRJNA236426). This data set consists of 110 individuals sequenced with the ABI SOLiD platform. Twenty of these samples belong to the C group, 30 belong to the M group, 30 belong to the A group, 20 belong to the O group and ten are A. cerana from Japan, which we used as an outgroup.
The 40 samples from Harpur et al. (2014) were all sequenced to high coverage, at an average of 38x per individual, which facilitated SNP calling and accurate calling of heterozygotes. The 110 samples from Wallberg et al. (2014) were primarily sequenced to low coverage, ∼4–6× mean sequence coverage, although four samples were sequenced to higher coverage using the SOLiD WildFire technology. All individuals from Harpur et al. (2014) and Wallberg et al. (2014) were diploid females. In addition, the Wallberg et al. (2014) data included one haploid male sequenced to 20× coverage. This was used to identify regions of the genome where inaccurate assembly may have produced incorrect inferences.
We also included six unpublished A. mellifera scutellata females from Kenya collected by Stephen Sheppard and sequenced on an Illumina platform to a mean coverage of between 11× and 27× per individual. These individuals are deposited in the Sequence Read Archive: PRJNA294105.
Alignments and SNP Calling
All of the Illumina-sequenced individuals from Harpur et al. (2014) and the six samples from Kenya were aligned with Bowtie2 using the very-sensitive-local alignment parameters (Langmead and Salzberg 2012). All of the SOLiD-sequenced individuals from Wallberg et al. (2014) were aligned using SHRiMP; an aligner specifically designed for colorspace sequencing data (Rumble et al. 2009). Different alignment procedures were used because of the differences in the format of the raw sequence data from the Illumina and SOLiD platforms. All data were aligned to the A . mellifera reference genome version 4.5 (www.beebase.org; last accessed April 2014).
We used the SAMtools/BCFtools packages (Li et al. 2009) to call genotypes, using a quality score threshold of 30 for the Harpur et al. (2014) data. We imposed some additional quality control filtering of these calls and kept only sites where the coverage for that individual was at least 10x, and we called heterozygous SNPs in individuals where we observed an alternate call at least twice. We used the SAMtools package (Li et al. 2009) to generate mpileup files from the Wallberg et al. (2014) data, using a quality score threshold of 250, and a custom Perl script to call genotypes. This alternate approach was used because SOLiD generates quality scores differently than Illumina. We kept all sites where the coverage for that individual was at least 5× and we called heterozygous SNPs in individuals where we observed an alternate call at least twice. We found that this strict calling approach was necessary in particular for the Wallberg et al. (2014) data set, where the error rate was high (further details below).
Additionally, we identified all heterozygous sites in the haploid male from Wallberg et al. (2014), for which we required that the minor allele represent at least 5% of the calls for a given site to consider the site biallelic. Sites that were identified as heterozygous in this sample were excluded from further analysis under the assumption that these sites were heterozygous due to errors in genome assembly. We identified 2,759,184 such sites over the entire 246.927 Mb genome.
We identified all sites in both A. mellifera and A. cerana samples that were variant in any individual, and then made genotype calls for each individual at these sites. We also counted the number and type of variant calls for each site. Within the Wallberg et al. (2014) data set, considering only A. mellifera individuals, we identified a total of 10,692,166 variable sites in at least one individual. Of these, 875,728 were identified as having two or more alternate SNP calls. Additionally, within single diploid individuals we found 328,194 sites with three or four alleles reported at that position or two or more non-reference allele calls with at least two observations of each base called. Within the Harpur et al. (2014) data set, considering only A. mellifera individuals, we identified 6,281,404 variant sites. Of these sites, 77 had two or more alternate SNP calls. The rate of triallelic calls in the Harpur et al. (2014) data set, ∼0.001% of variant sites, is much more consistent with 0.083–1.86% of variant sites that are triallelic as observed in Drosophila and hominid genomes (Sepylarskiy et al. 2012). This figure contrasts with the 8.2% of variant sites that are triallelic in the Wallberg et al. (2014) data set. While we expect a certain level of triallelic SNPs to be truly segregating within A. mellifera populations, such high rate of triallelic alleles likely reflects an elevated sequencing error rate associated with SOLiD sequencing.
To further examine the potential differences in error rate between sequencing platforms, we examined the relationship between mean coverage and the rate at which a heterozygote call was observed (supplementary tables S1 and S2, Supplementary Material online). In the Wallberg et al. (2014) data set, we observed a strong and significant correlation between mean coverage and heterozygosity (R2 = 0.88; P < 2e-16). We found this to be true even when we excluded the four high coverage individuals sequenced using the WildFire method (R2 = 0.3022; P = 6.252e-10). In contrast, we found no such relationship in the Harpur et al. (2014) data set (R2 = −0.0207; P = 0.6348). This could be a true reflection of higher coverage allowing for more accurate detection of heterozygotes, or it could be a signal of sequencing error. To further test this, we regressed the number of triallelic positions called for an individual diploid bee against sequencing coverage for the Wallberg et al. (2014) data set. We found that, as coverage increased, the number of triallelic sites called within an individual also increased (R2 =0.83; P < 2.2e-16). This pattern remained even when we excluded the four high coverage individuals (R2 = 0.41; P < 2.89 e-12), further indicating that most triallelic positions and many heterozygous sites in the Wallberg et al. (2014) data set are likely generated via sequencing error by the SOLiD platform. We found that the four samples sequenced to higher coverage contributed 93.6% of the triallelic positions. Therefore, we excluded these samples when selecting SNPs for downstream analysis. We included these samples in our subsequent analyses, but sites within these samples were included only if we found a SNP at the same site in another individual. This means that we are excluding variants private to one of these four samples due to our inability to differentiate true variants from error.
SNP Selection
We refined our data set to include only sites that were genotyped for the majority of individuals in across all A. mellifera populations. For a site to be included in our downstream analysis we required each site to have sufficient coverage to make a genotype call in at least 80/96 individuals from the Wallberg et al. (2014) data set and in at least 36/39 individuals from the Harpur et al. data set. The number 96 reflects that we excluded the 4 samples that were sequenced to high coverage in the Wallberg et al. (2014) data set. We then added to this data set additional sites called in at least 8/10 A. cerana individuals from the Wallberg et al. (2014) data set as well as the one A. cerana from Harpur et al. (2014).
Our initial analysis of the combined data set indicated the possibility of spurious results. For instance, all Harpur et al. (2014) individuals grouped together in an ADMIXTURE analysis and exhibited no population differentiation in a PCA analysis (data not shown). We therefore attempted to correct for residual sequencing errors in two ways. Because the error rate for base calling is substantially lower for the Illumina sequence data than for the SOLiD sequencing data, whenever we had information about a SNP from the Illumina data we only accepted information about that site in a SOLiD sequenced individual when the SOLiD data either (1) indicated the reference base or (2) when it indicated a SNP that had been observed in the Illumina data. Therefore at sites where we had information from Illumina sequencing, we were able to filter out SNPs likely due to SOLiD sequencing error. However, to avoid biasing our information by only including those SNPs observed in the Harpur et al. (2014) data set, we also included additional SNPs from the Wallberg et al. (2014) data if only one alternate SNP was observed at that site over all individuals. This is particularly important for the O group and the A. cerana populations where there were either zero or one individuals from the Illumina data, respectively. While this method will not be able to completely remove all errors from the data set, we balance this with avoiding bias in calling SNPs in different populations. This procedure yielded a set of 832,654 sites that passed our coverage and inclusion cutoffs and were therefore used in all downstream analyses.
Tests of Population Structure
We used the program Dadi (Gutenkunst et al. 2009) to examine whole genome patterns of differentiation between population pairs. FST values for population pairs as well as joint allele frequency spectra were generated to assess differentiation between populations (supplementary fig. S1, Supplementary Material online). For our initial exploration of potential admixture within the entire set of A. mellifera individuals we ran ADMIXTURE (Alexander et al. 2009) with K values 2 through 7 using the cross validation procedure and the -s time option to set the random seed using the clock time.
To examine the patterns of relatedness between individuals and among major groups, we created a distance matrix from the genotype data using SNPRelate (Zheng et al. 2012). We generated multiple matrices based on different subsets of the data that correspond to the samples used in the Whitfield et al. (2006) and the Wallberg et al. (2014) studies. We then ran a non-parametric multidimensional scaling analysis (nMDS) using the R package ecodist version 1.2.2 and calculated the stress value and R2 value for each analysis. We ran the adonis function from the vegan version 2.3-4 package in R to identify the effect of major groupings on the distance matrix.
We used TreeMix version 1.12 (Pickrell and Pritchard 2012) to generate a maximum likelihood tree using all populations. We repeated the analysis using the Harpur et al. (2014) data and the Kenyan samples sequenced on the Illumina platform separately. We further tested adding migration events to the resulting TreeMix trees for each analysis.
The maximum likelihood tree was then used as the correct model for formal tests of admixture using ADMIXTOOLS (Patterson et al. 2012). We calculated f3 statistics for all pairs of source populations for each potential target population to identify populations with evidence of admixture. We then kept all results for which we found evidence of admixture with an f3 statistic less than −0.01 and a z-score at least four standard deviations from the mean. For each f3 statistic that passed our filtering cutoffs we calculated both D statistics, to determine whether pairs of populations form a clade or if there is evidence of gene flow between the hypothesized population pairs. F4 ratio estimation was performed on the same set of f3 statistics that passed our filters to estimate the proportion of ancestry in the admixed populations.
Geographic Differentiation
We first examined population level differences between all pairs of major honeybee lineages by combining all individuals from each group into a single population. We calculated FST for each site for each population pair where we had genotype information for at least 90% of individuals in each population in the pair for that site. We identified for each site in each population pair if the site was in an exonic region and if so, if the SNP difference produces a synonymous or non-synonymous amino acid. We also identified the 99th percentile of FST differences for each population pair and identified the list of genes with at least one SNP in this category.
For each potential pair of major A. mellifera groups we compared the proportion of SNPs in the 99th percentile of FST differences for exonic and intronic regions with the genome wide proportion. We used a Fisher’s exact test to identify differences in observed versus expected proportions of exonic and intronic SNPs between population pairs. For each site identified in an exon, we also identified which amino acid it coded for and the alternate amino acid encoded for by the alternate base.
Gene Ontology Analysis
We used the DAVID Functional Annotation tool (Huang et al. 2009a, 2009b) to identify enriched gene ontology terms for each pair of populations. Our gene lists were limited to those genes with ortholog information for Drosophila melanogaster downloaded from the National Center for Biotechnology Information (ncbi.nlm.nih.gov), as this information is used to connect gene identifiers to gene ontology terms. We used the medium stringency for gene classification in DAVID and used an enrichment score of 1.3 as the minimum score for identifying enriched clusters and the Benjamini corrected P-value (Benjamini and Hochberg 1995) to identify significantly enriched terms within clusters.
Results
We identified 832,654 sites across the A. mellifera nuclear genome, following screening (see Materials and Methods for details), where a SNP was identified in one or more individuals and where we were also able to genotype most individuals at that site. The majority of SNPs identified were private to a single major lineage, with the A group containing the largest number of private SNPs (table 1). We calculated mean π, nucleotide diversity, in 10Kb windows for each major lineage of A. mellifera, as well as by population, from each data set (table 2). We found that the A group (African populations) exhibited the highest mean π values, an observation consistent with previous findings (Whitfield et al. 2006; Wallberg et al. 2014). The O and Y lineages had mean π values intermediate to the A lineage and the C and M lineages.
Table 1.
Number of Private SNPs by Major Lineage
| Group | Number of SNPs | Number of Individuals in Group |
|---|---|---|
| A. cerana | 99178 | 10 |
| O | 53372 | 20 |
| Y | 43561 | 10 |
| C | 37660 | 29 |
| M | 72809 | 39 |
| A | 362518 | 47 |
| Total | 669098 | 155 |
Table 2.
Mean π in 10-kb Overlapping Windows with a 1-kb Slide
| Group | Mean π (10-kb windows) | N |
|---|---|---|
| A | 1.65E-03 | 47 |
| C | 5.26E-04 | 29 |
| M | 8.47E-04 | 39 |
| O | 1.17E-03 | 20 |
| Y | 1.05E-03 | 10 |
| Kenya (A) | 1.49E-03 | 6 |
| Harpur et al. 2014 | ||
| A | 1.83E-03 | 11 |
| C | 4.65E-04 | 9 |
| M | 7.34E-04 | 9 |
| Y | 1.05E-03 | 10 |
| Wallberg et al. 2014 | ||
| Austria (C) | 6.38E-04 | 10 |
| Italy (C) | 4.78E-04 | 10 |
| Jordan (O) | 1.35E-03 | 10 |
| Nigeria (A) | 1.70E-03 | 10 |
| Norway (M) | 8.56E-04 | 10 |
| South Africa: capensis (A) | 1.68E-03 | 10 |
| South Africa: scutellata (A) | 1.69E-03 | 10 |
| Spain (M) | 9.81E-04 | 10 |
| Sweden (M) | 7.60E-04 | 10 |
| Turkey (O) | 8.83E-04 | 10 |
FST values between pairs of major genetic groups showed the highest differentiation between the Y group and all other lineages (table 3). This observation is consistent with the initial analysis of this population by Franck et al. (2001). The differentiation between the A group and the each of the C, M, and O groups was lower than the pairwise differentiation among the C, M, and O groups, suggesting that these populations are more closely related to the A group, which is consistent with the hypothesis of A being the population source for these lineages (Whitfield et al. 2006). The O group is also more differentiated from the Y group than it is from the A group. We generated joint site frequency spectra with Dadi for each pair of populations (supplementary fig. S1, Supplementary Material online). We find that differences between the M lineage and the C lineage are most clearly pronounced with gaps in the joint frequency spectra at intermediate and high frequency SNPs in both populations. This is consistent with these two lineages representing separate colonization events of Europe even though they are geographically adjacent (Whitfield et al. 2006; Han et al. 2012).
Table 3.
FST Values for Pairs of Populations
| A | C | M | O | Y | |
|---|---|---|---|---|---|
| A | — | ||||
| C | 0.134 | — | |||
| M | 0.191 | 0.268 | — | ||
| O | 0.134 | 0.237 | 0.308 | — | |
| Y | 0.252 | 0.423 | 0.410 | 0.354 | — |
Ancestry Estimation in A. m ellifera Individuals
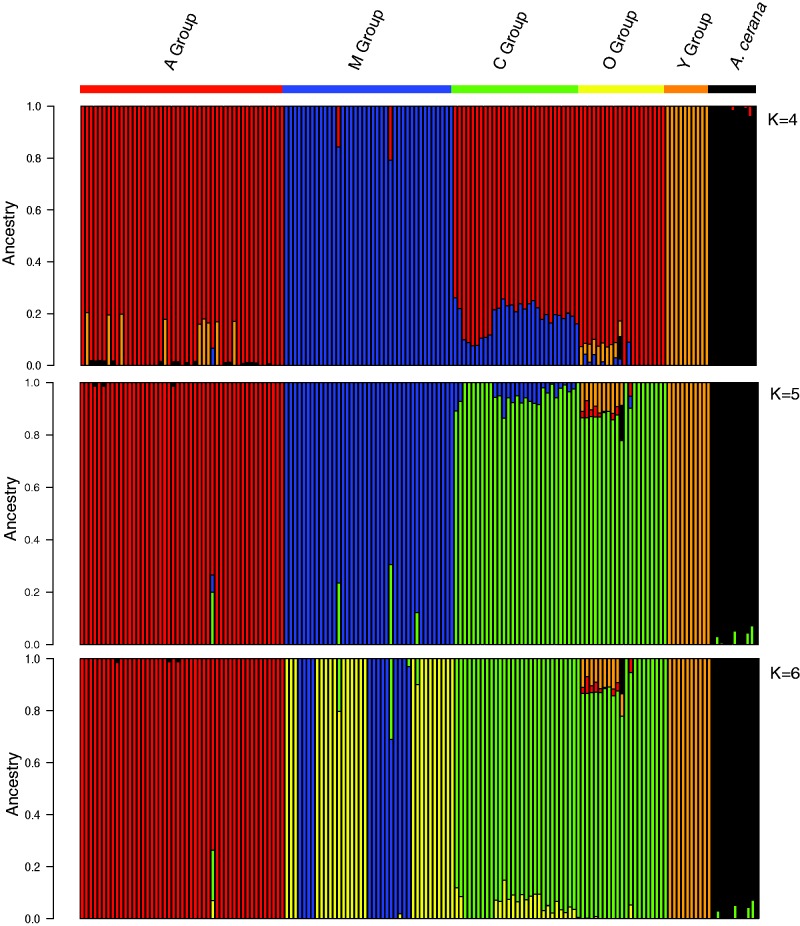
We investigated the partitioning of individuals into clusters to identify potentially admixed populations using the program ADMIXTURE (Alexander et al. 2009) (fig. 2, supplementary fig. S2, Supplementary Material online). The overall clustering of individuals was consistent with the major lineages identified in previous studies. When we ran the analyses setting the value of K to 6 or less, the C and O lineages clustered together, suggesting that these lineages are less differentiated from each other than they are from other groups, and supporting the hypothesis that the C lineage and O lineage share a common ancestor (Whitfield et al. 2006; Wallberg et al. 2014). The C and O lineages are in geographic proximity to one another, with C individuals sampled from Italy and Austria and O individuals sampled from Turkey and Jordan (fig. 2). Cross validation procedures indicated that a K of 5 was optimal. Previous analyses of these lineages also support this clustering (Whitfield et al. 2006; Wallberg et al. 2014). The M lineage splits at K = 6 into two distinct groups, one including all individuals from Spain and the other including all individuals from Northern Europe—these groups include individuals from both data sets.
Fig. 2.—
ADMIXTURE results for K values of 4 to 6. The Jordanian population shows evidence of substantial admixture between C/O populations and the Y group.
A surprising result was that the Middle Eastern Y lineage individuals cluster together with African individuals at K values of 3 or less and then become distinct at K = 4 (fig. 2). However, the Y lineage does not cluster with individuals from the Middle Eastern O lineage at any K value despite the geographic proximity of Y and O populations. Moreover, the Jordanian population appears to be admixed, receiving contributions from both Y and C/O lineages at K = 4 through 6, further demarcating the separation between the O and Y groups (fig. 2)
One individual from Jordan consistently showed ancestry as derived from A. cerana in the ADMIXTURE results. We believe that this is an unlikely biologically scenario (given that these two species diverged 6–25 million years ago) but instead, we suggest that this pattern has been produced by residual error in the Wallberg et al. (2014) data set (see Materials and Methods and supplementary table S1, Supplementary Material online).
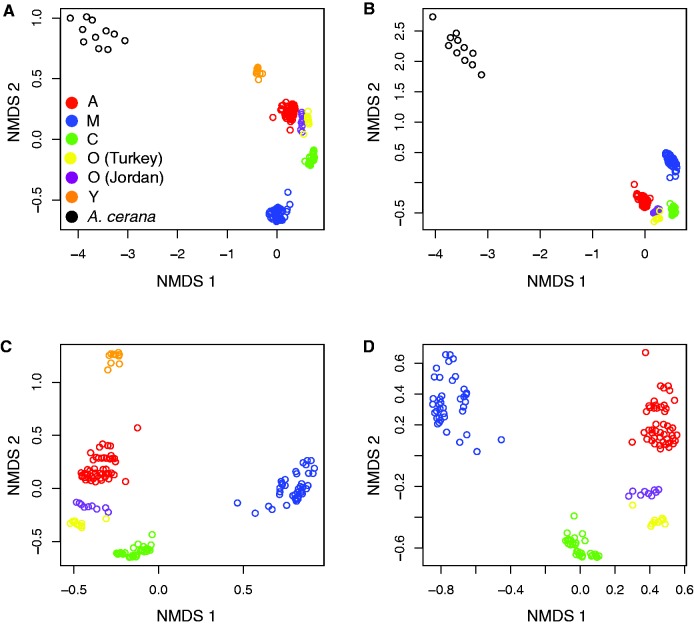
We generated a dissimilarity matrix based on identity by state at each site for each pair of individuals using SNPRelate (Zheng et al. 2012) and performed a series of non-metric multidimensional scaling (nMDS) analyses on subsets of the data comparable to the data available in previous studies (Whitfield et al. 2006; Wallberg et al. 2014). When we plotted the pairwise distances between all individuals in the data set, the Y group individuals were placed in a two-dimensional space between the A. cerana population and the A group (fig. 3A; R2 = 0.998, stress = 0.0323) indicating a good fit of the data to the model (supplementary table S3, Supplementary Material online). This pattern is consistent with the hypothesis that either northwestern Africa or the Middle East represent the likely centers of origin of A. mellifera, as previously proposed based on the observation of multiple mitochondrial lineages in this region (Ruttner et al. 1978; Franck et al. 2001) as the Y and A individuals are located most closely to the A. cerana outgroup (fig. 3A ).
Fig. 3.—
Non-parametric multidimensional scaling analyses on a dissimilarity matrix summarizing kinship data between pairs of individuals for (A) All individuals, (B) Y group removed, an analysis directly comparable to Wallberg et al. (2014), (C) A. cerana removed and (D) Y group and A. cerana removed, an analysis directly comparable to the Whitfield et al. (2006) analysis.
Further exploration of subsets of the data indicated consistent groupings based on the pairwise distances between individuals. Removal of the Y population (fig. 3B ) did not produce any difference in the distances between remaining individuals. This analysis is directly comparable to that done by Wallberg et al. (2014) as it included individuals from the same major lineages.
Further examinations including the removal of the A. cerana population (fig. 3C ) or both the A. cerana and Y group populations (fig. 3D ) did not alter the pairwise distances between remaining individuals. Figure 3D is directly comparable to the sets of populations used in Whitfield et al. 2006 and agrees with the results of the PCA analysis (fig. 1A ) in that study. In each case, we find that the data fit the model well (supplementary table S3, Supplementary Material online). These data are consistent with the hypothesis that the M, C, and O lineages are derived from an ancestral African population, as proposed by Whitfield et al. (2006). In particular, the distances between individuals in figure 3A , where all populations are considered, are more consistent with an origin of A. mellifera in northwestern Africa or the Middle East than with an ancient split between the A and O lineages.
Additional model fitting in three dimensions produced marginally better fits of the model to the data (supplementary table S3, Supplementary Material online). We calculated the effect of group on the variation in each subset of the data, using the R package adonis, and found a significant effect in all cases (P = 0.001 for each comparison). Taken together with the clustering results in the ADMIXTURE analysis, these results support the idea that C and O lineages represent one migration out of Africa with subsequent diversification. The placement of the O lineage between the C and A lineages indicates that the A and O lineages likely share a common ancestor more distantly than C and O lineages. The placement of the Y group between A. cerana and the rest of A. mellifera in the nMDS analysis and the clustering of the Y and A groups at low K values in the ADMIXTURE analysis indicate that the Y and A lineages are likely derived from a population ancestral to the rest of A. mellifera.
Geographic Differentiation within A and M Groups
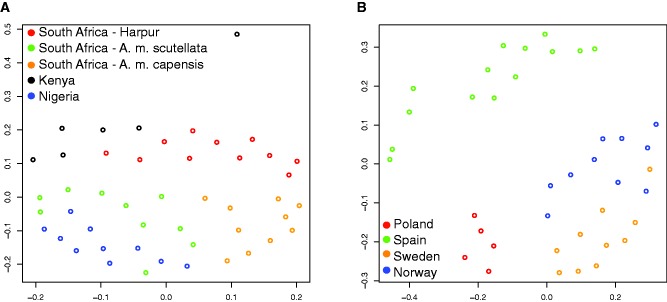
A total of 47 A group individuals were included in the combined data set, including six A. m. scutellata from Kenya, ten A. m. adansonii from Nigeria, and 31 from South Africa. Within the South African individuals, there were 11 from Harpur et al. (2014) and 20 from Wallberg et al. (2014), ten of which were identified as A. m. capensis and ten identified as A. mellifera scutellata. Because we did not see clustering of A group individuals based on geography in our ADMIXTURE analysis, despite our expectation of differences between A group subspecies, we ran an nMDS analysis on the A group individuals alone, following the same procedures indicated above, to determine whether any geographic distinctions emerged when the relatedness between these individuals were considered separately. For comparison, we ran the same sets of analyses on the M group individuals. The M group was chosen because the A and M groups have the most similar number of individuals and number of geographic locations from which individuals were drawn: 14 A. m. iberiensis from Spain, and A. m. mellifera individuals; ten from Norway, ten from Sweden, and five from Poland.
The nMDS analysis of all 47 A group individuals exhibited little differentiation among individuals that reflected geographic distances (fig. 4A ); however, the data did not exhibit a good fit to the model (supplementary table S3, Supplementary Material online). We explored higher numbers of dimensions, but the fit of the data improved only slightly. The control analysis of M group individuals partitioned the M group into two distinct subsets, Spain vs. Northern Europe, based on geography (fig. 4B ), but the fit of the data to the model is only slightly better than for the A group. This is similar to the splitting of individuals from Spain vs. Northern Europe in the ADMIXTURE analysis (fig. 2). We therefore consider it unlikely that methodological or systemic differences in the data sets that are responsible for the patterns reported here, but rather, suggest that the observed patterns reflect a biological phenomenon. Both the A and the M groups appear to be best described as large panmictic groups with slight geographic differentiation in the M group between Spain and Northern Europe. However, the overall picture is consistent with the idea that there is substantial gene flow throughout these groups as gene flow would represent the simplest explanation for lack of geographic differentiation between individuals within lineages.
Fig. 4.—
Non-parametric multidimensional scaling analyses on an dissimilarity matrix summarizing kinship data between pairs of individuals for (A) All A group individuals, (B) All M group individuals.
Tests of Admixture
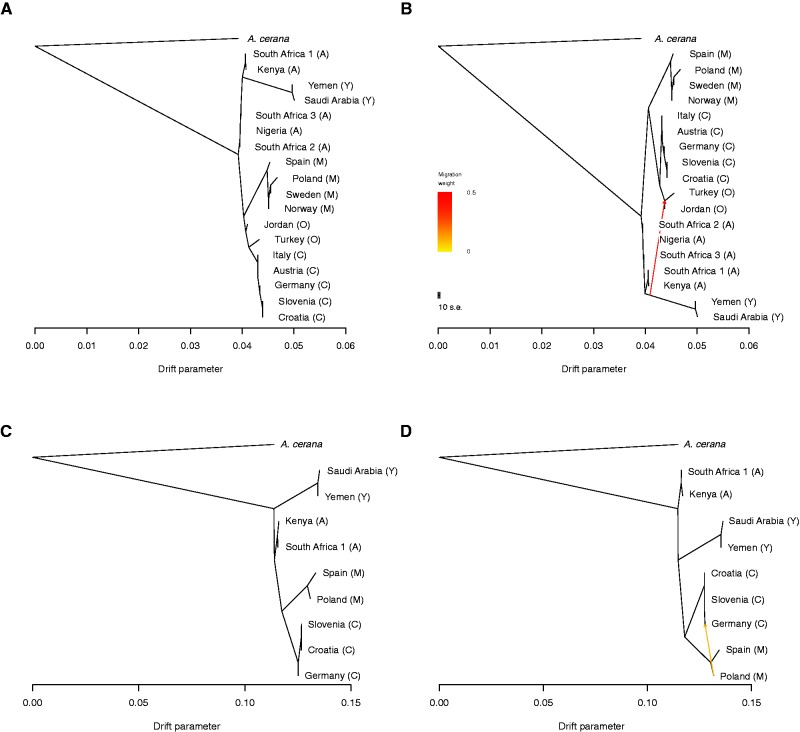
To test for signatures of recent admixture among populations, we generated a maximum likelihood tree using the TreeMix approach (Pickrell and Pritchard 2012). We first generated a tree with no migration events (fig. 5A ) including all populations. The maximum likelihood tree places the root of A. mellifera between the Y and A lineages and the group leading to the C, O, and M lineages. The tree also places the O lineage populations adjacent to the C lineage population. Adding a migration event to this tree does not substantively change the conformation of the tree, but does indicate a strong migration event from Y to the base of the O group (migration weight 46%; P <2.22507e-308) (fig. 5B ).
Fig. 5.—
(A) TreeMix tree with no migration events, including all populations (B) TreeMix tree with one migration event, including all populations (C) TreeMix tree with no migration events including only Illumina sequenced data and (D) TreeMix tree with one migration event including only Illumina sequenced data.
We also generated a maximum likelihood tree for the subset of individuals that were sequenced with Illumina only. This tree has some differences with respect to the tree that included all individuals and populations. In particular, the most basal node within the A. mellifera split was the Y group and the A, C, and M groups (fig. 5C ). The A group is placed intermediate to the basal node and the node that splits the C and M groups. Adding a single migration event adds a migration from Poland in the M group populations to the C group German population (migration weight 10%; P <2.22507e-308, fig. 5D ). We then used the maximum likelihood tree topology shown in figure 5A as the basis for further tests of admixture.
We performed three population tests of admixture, calculating the f3 statistic, using ADMIXTOOLS (Patterson et al. 2012). We tested for possible admixture in all populations considering all other pairs of populations as potential population sources. Evidence of admixture is indicated by a negative f3 statistic. For tests where we identified a negative f3 statistic, we calculated the upper and lower bound on the admixture proportion (table 4). Using this approach, we retained results for the populations with the most negative f3 statistics: f3 ≤ -0.02. We found evidence of admixture in the central European populations from Austria, Germany and Italy, all of which are C group populations. These populations all display evidence of admixture when the source populations are one of two eastern European C group populations, Slovenia and Croatia, and an M group population. The z-scores for tests between the C group and M group populations are larger than those obtained between the C group and non-European populations suggesting that non-European populations, which are geographically more distant from the target populations, are likely instances of the outgroup case and reflect more ancient relationships between the populations. We also calculated D statistics as a test of clades for each of these sets of populations and found consistent results (supplementary table S4, Supplementary Material online).
Table 4.
The f3 Tests Showing Evidence of Admixture with an f3 ≤ −0.02 as well as the Upper and Lower Bound on the Mixing Proportions
| Source 1 | Source 2 | Target | F3 | Z-score | αL | αU |
|---|---|---|---|---|---|---|
| Croatia | Norway | Austria | −0.062192 | −34.487 | 0.798 | 0.883 |
| Croatia | Sweden | Austria | −0.06073 | −33.093 | 0.795 | 0.89 |
| Croatia | Poland | Austria | −0.060628 | −29.318 | 0.805 | 0.917 |
| Croatia | Spain | Austria | −0.057976 | −32.241 | 0.811 | 0.897 |
| Slovenia | Norway | Austria | −0.05559 | −28.203 | 0.797 | 0.897 |
| Slovenia | Sweden | Austria | −0.053905 | −27.189 | 0.793 | 0.905 |
| Slovenia | Poland | Austria | −0.053336 | −23.684 | 0.803 | 0.93 |
| Slovenia | Spain | Austria | −0.052037 | −26.207 | 0.81 | 0.911 |
| Norway | Slovenia | Germany | −0.06205 | −18.595 | 0.097 | 0.107 |
| Poland | Croatia | Germany | −0.081838 | −23.03 | 0.101 | 0.105 |
| Poland | Slovenia | Germany | −0.074376 | −19.566 | 0.089 | 0.105 |
| SaudiArabia | Slovenia | Germany | −0.020572 | −8.555 | 0.03 | 0.075 |
| Spain | Slovenia | Germany | −0.062854 | −18.555 | 0.092 | 0.1 |
| Sweden | Slovenia | Germany | −0.062873 | −18.591 | 0.095 | 0.108 |
| Yemen | Slovenia | Germany | −0.020819 | −8.667 | 0.031 | 0.076 |
| Croatia | Norway | Italy | −0.063827 | −32.188 | 0.757 | 0.9 |
| Croatia | Spain | Italy | −0.063037 | −32.213 | 0.772 | 0.91 |
| Croatia | Sweden | Italy | −0.062479 | −31.449 | 0.753 | 0.898 |
| Croatia | Poland | Italy | −0.053326 | −24.067 | 0.765 | 0.909 |
| Croatia | Nigeria | Italy | −0.028435 | −18.775 | 0.808 | 0.935 |
| Slovenia | Norway | Italy | −0.056161 | −26.716 | 0.754 | 0.891 |
| Slovenia | Spain | Italy | −0.056102 | −26.727 | 0.771 | 0.901 |
| Slovenia | Sweden | Italy | −0.05452 | −26.006 | 0.751 | 0.888 |
| Slovenia | Poland | Italy | −0.044811 | −18.943 | 0.762 | 0.899 |
| Slovenia | Nigeria | Italy | −0.031338 | −21.195 | 0.81 | 0.928 |
| Slovenia | Yemen | Italy | −0.021996 | −12.267 | 0.828 | 0.936 |
| Slovenia | SaudiArabia | Italy | −0.020555 | −11.689 | 0.828 | 0.937 |
| SouthAfrica | Slovenia | Italy | −0.027977 | −18.294 | 0.068 | 0.183 |
| SouthAfrica | Croatia | Italy | −0.02505 | −16.127 | 0.062 | 0.185 |
| SaudiArabia | Turkey | Jordan | −0.032375 | −32.705 | 0.066 | 0.498 |
| Yemen | Turkey | Jordan | −0.031437 | −31.485 | 0.068 | 0.498 |
| Croatia | Poland | Norway | −0.030838 | −12.903 | 0.059 | 0.212 |
| Germany | Poland | Norway | −0.022496 | −10.022 | 0.066 | 0.236 |
| Poland | Italy | Norway | −0.036531 | −16.71 | 0.781 | 0.912 |
| Poland | Austria | Norway | −0.031329 | −14.967 | 0.769 | 0.934 |
| Poland | Jordan | Norway | −0.02429 | −12.238 | 0.799 | 0.946 |
| Poland | Nigeria | Norway | −0.021525 | −12.664 | 0.809 | 0.95 |
| Slovenia | Poland | Norway | −0.030407 | −12.814 | 0.06 | 0.215 |
| Croatia | Poland | Sweden | −0.022538 | −8.382 | 0.047 | 0.204 |
| Poland | Italy | Sweden | −0.027648 | −11.074 | 0.79 | 0.894 |
| Poland | Austria | Sweden | −0.021911 | −9.229 | 0.778 | 0.928 |
| Slovenia | Poland | Sweden | −0.022201 | −8.344 | 0.046 | 0.206 |
We found evidence of admixture in two populations from the M group (Sweden and Norway) as well as admixture in the Jordanian population. The Jordanian population shows evidence of admixture between Turkey, an O group population, and the Y group populations as suggested by the ADMIXTURE results, the TreeMix analysis and our nMDS analysis. We do not find evidence of admixture in any of the African populations.
We further calculated the F4 ratio to estimate ancestry proportions for our identified admixed populations (table 5). We found that the proportion of admixture contributed by northern and western European populations to the C group populations from Germany and Austria, is 5 - 15% depending upon the source population considered. The M group populations from Norway and Sweden show 25–36% ancestry from central European populations. The Jordanian population derives 35% of its ancestry from Y group populations and the remaining 65% from the Turkish, O group, population.
Table 5.
The F4 Ratio Statistic for the Set of f3 Tests Showing Evidence of Admixture in the Target Population
| Outgroup | A | B | C | X | α | SE | Z Score |
|---|---|---|---|---|---|---|---|
| A. cerana | SouthAfrica | Norway | Croatia | Austria | 0.060884 | 0.047895 | 1.271 |
| A. cerana | SouthAfrica | Sweden | Croatia | Austria | 0.059568 | 0.045241 | 1.317 |
| A. cerana | SouthAfrica | Spain | Croatia | Austria | 0.04974 | 0.038152 | 1.304 |
| A. cerana | SouthAfrica | Norway | Slovenia | Austria | 0.091662 | 0.045646 | 2.008 |
| A. cerana | SouthAfrica | Sweden | Slovenia | Austria | 0.08863 | 0.04343 | 2.041 |
| A. cerana | SouthAfrica | Poland | Slovenia | Austria | 0.05203 | 0.032832 | 1.585 |
| A. cerana | SouthAfrica | Spain | Slovenia | Austria | 0.073918 | 0.037033 | 1.996 |
| A. cerana | SouthAfrica | Slovenia | Norway | Germany | 0.850214 | 0.055054 | 15.443 |
| A. cerana | SouthAfrica | Croatia | Poland | Germany | 0.920308 | 0.04035 | 22.808 |
| A. cerana | SouthAfrica | Slovenia | Poland | Germany | 0.895274 | 0.039268 | 22.799 |
| A. cerana | SouthAfrica | Slovenia | SaudiArabia | Germany | 0.926164 | 0.029394 | 31.508 |
| A. cerana | SouthAfrica | Slovenia | Spain | Germany | 0.881468 | 0.043644 | 20.197 |
| A. cerana | SouthAfrica | Slovenia | Sweden | Germany | 0.857545 | 0.052073 | 16.468 |
| A. cerana | SouthAfrica | Slovenia | Yemen | Germany | 0.928155 | 0.030037 | 30.901 |
| A. cerana | SouthAfrica | Turkey | SaudiArabia | Jordan | 0.652508 | 0.071216 | 9.162 |
| A. cerana | SouthAfrica | Turkey | Yemen | Jordan | 0.652395 | 0.073125 | 8.922 |
| A. cerana | SouthAfrica | Poland | Croatia | Norway | 0.673078 | 0.044174 | 15.237 |
| A. cerana | SouthAfrica | Poland | Germany | Norway | 0.640784 | 0.047591 | 13.464 |
| A. cerana | SouthAfrica | Italy | Poland | Norway | 0.300607 | 0.036469 | 8.243 |
| A. cerana | SouthAfrica | Austria | Poland | Norway | 0.340266 | 0.04255 | 7.997 |
| A. cerana | SouthAfrica | Poland | Slovenia | Norway | 0.679728 | 0.042605 | 15.954 |
| A. cerana | SouthAfrica | Poland | Croatia | Sweden | 0.714591 | 0.042468 | 16.827 |
| A. cerana | SouthAfrica | Italy | Poland | Sweden | 0.259854 | 0.036072 | 7.204 |
| A. cerana | SouthAfrica | Austria | Poland | Sweden | 0.292335 | 0.041754 | 7.001 |
| A. cerana | SouthAfrica | Poland | Slovenia | Sweden | 0.719136 | 0.040962 | 17.556 |
Note.—The alpha value shows the proportion of ancestry from the underlined population to the italicized population.
Geographic Differentiation
For each pair of major groups we calculated the 95th percentile for FST for each site for which we had sufficient coverage at the site in each population. We found that, within this set of differentiated SNPs, there were more SNPs in exonic regions than expected between every population pair, given the null expectation that SNPs will be randomly located throughout the genome (supplementary table S5, Supplementary Material online). In A. mellifera, the exonic regions made up 13.4% of the genome, but accounted for 16.1–23.5% of SNPs between population pairs. Similarly, while the intronic regions made up 43.9% of the genome, they accounted for 37.4–41.1% of SNPs between population pairs (supplementary table S6, Supplementary Material online), a significant difference, though not as extreme as the difference in exonic sequence. We also found slightly fewer than expected SNPs within intergenic regions in a couple of population pairs, but no difference from the expectation in most comparisons (supplementary table S7, Supplementary Material online). Intergenic sequence accounted for 42.7% of the genome and we found 39.1–42.9% of SNPs in this category.
Within the set of exonic differentiated SNPs, we found dozens of non-synonymous SNPs between all population pairs. We calculated the number of non-synonymous SNPs per kilobase of exon for each gene for each population pair where we identified one or more differentiated SNPs within that gene. We found substantial non-synonymous SNP variation in the gene dumpy; which is important for epidermal-cuticle attachment during morphogenesis (Wilkin et al. 2000) and may contribute to morphological differences observed between A. mellifera populations. In dumpy we found 12 differentiated non-synonymous SNPs between the A and Y lineages, which translates to 0.21 non-synonymous SNPs per KB of exon. We also found differentiated non-synonymous SNPs in well-studied A. mellifera genes such as vitellogenin, which is involved in caste determination (Engels et al. 1990), and the major royal jelly protein genes (Drapeau et al. 2006), which are involved in reproductive maturation. In vitellogenin we found differentiated non-synoymous SNPs between every population pair, ranging from 0.1844 SNPs/Kb and 0.369 SNPs/Kb of exon.
Gene Ontology Analysis
We used DAVID version 6.8 (Huang et al. 2009a, 2009b), to identify gene ontology (GO) categories that are enriched in sets of genes with one or more SNP in the ≥ 95th percentile category for each population pair. Enriched clusters of GO terms were found in all of the population pair comparisons except between the M and C lineages.
We found enrichment in the category of transmembrane, transmembrane helix, and membrane associated with a variety of cellular activities in every comparison between population pairs where we found enriched clusters (supplementary tables S8–16, Supplementary Material online). We observed non-synonymous variation in 94/724 (13%) of SNPs within 294 genes associated with these terms. Sensory related terms were enriched in many of the comparisons between groups and 37/318 (11.6%) SNPs found in genes associated with these terms were non-synonymous.
Discussion
The relationships among populations of A. mellifera have historically been difficult to disentangle, and the origin of this species has been the subject of substantial debate (Sheppard and Meixner 2003; Whitfield et al. 2006; Han et al. 2012; Wallberg et al. 2014). One major hurdle to understanding the demographic and evolutionary history of honey bees is the existence of several genetically distinct populations in close geographic proximity in the Middle East, Africa, and Western Asia. Much of the current literature on this topic has been focused on two of these lineages: the A lineage and the O lineage. A 2006 analysis of the A, C, M, and O lineages concluded that the M, C, and O lineages are derived from Africa (Whitfield et al. 2006). The results of a more recent study (Wallberg et al. 2014) were most consistent with a previous hypothesis placing the geographic origin of A. mellifera in the Middle East, with the A and O linages both deriving from an original population (see Han et al. 2012; fig. 1B i for a pictorial representation of this hypothesis). Our analysis applied the largest currently available set of A. mellifera sequence data to this issue, and revealed that A. mellifera has a complex demographic history beyond previous findings. Our analysis, unlike previous studies, contains populations of the Y lineage with individuals from both Saudi Arabia and Yemen. These individuals provide key information about the origins of A. mellifera because they elucidate some of the confusing or unclear relationships among the major lineages in other studies, in particular, Han et al. (2012), Alburaki et al. (2013), and Wallberg et al. (2014).
We hypothesize an evolutionary history for A. mellifera in which the origin of A. mellifera is located in the Middle Eastern or northeastern African, having diverged from its nearest relatives in Asia. However, in contrast to earlier hypotheses that place the origin of A. mellifera in this region (Ruttner 1988; Han et al. 2012; Wallberg et al. 2014), we find that the M, C, and O lineages are derived from Africa instead of an ancient split between the A and O lineages. Our model proposes that, following the origin of A. mellifera, substantial diversification occurred in Africa followed by radiations of the M lineage out of Africa into Europe, and of the C/O lineages back into the Middle East, and then into central Europe leading to the modern European and western Asian populations. In addition, we find support that the Y lineage, which occurs in Northeastern Africa and the Arabian Peninsula (Franck et al. 2001; Harpur et al. 2014), is also derived from the basal A. mellifera population. The inclusion of the Y population in our analysis is key to clarifying many of the confusing and contradictory patterns that have been previously discussed in the literature. For instance, our analysis allowed us to identify patterns of relatedness between populations that would otherwise remain cryptic, such as admixture between the O lineage and the Y lineage in Jordan, a relationship that was previously misidentified as admixture between the O lineage and the A lineage (Wallberg et al. 2014). A similar situation likely occurred in the study of A. mellifera from Syria, by Alburaki et al. (2013). In that study, which relied on microsatellite data, A. mellifera from Syria were associated with both the A group and the O group in different analyses. This confusing placement can be explained, however, if these bee populations, like the geographically nearby Jordanian bee populations analyzed here, are not examples of a pure lineage, but instead, represent admixed individuals that derive ancestry from both from O lineage individuals (included in the Alburaki et al. 2013 analysis) and Y lineage individuals (which were not included). We suggest that the bees from Syria, like the bees from Jordan, are in a secondary contact zone between the Y and the O lineages and that further investigation including populations from this area would be informative.
Our analysis is based on the most comprehensive honey bee genomic dataset to date. By analyzing subsets of the data, our study reconciles the divergent conclusions that were reached by previous studies. In addition, our analysis illustrates how disentangling complex relationships between populations requires the appropriate population comparisons. When the entire data set is considered, our nMDS analysis places the Middle Eastern Y lineage between A. cerana from the rest of the A. mellifera populations (fig. 3A ). In this analysis, the Jordanian population is also recovered as more closely related to the Y population than the Turkish O population is to the Y population, supporting a scenario of admixture within Jordanian bee populations. This relationship is well supported in our analysis, with additional ADMIXTURE results indicating that the Jordanian populations is composed of a proportion of Y as well as C/O ancestry, and a confirmation by both f3 statistics and the F4 ratio analysis. However, the Wallberg et al. (2014) analysis did not identify this pattern correctly, and instead attributed ∼18% of the Jordanian populations ancestry to the African A group. This confusing pattern is easily understood by examining figure 3. The Jordanian population is closely related to the other O lineage as expected by its geographic proximity. However because both the A lineage and the Jordanian population are intermediate between the Y lineage and the O lineage (since O is also derived from Africa), these two populations are placed near each other. Thus, a more detailed analysis, such as the formal tests of admixture using ADMIXTOOLS that we conducted, can clarify the patterns of relatedness between these populations. In our data set, we find an F3 statistic of −0.032375 for Jordan when considering Turkey and Saudi Arabia as source populations. On the other hand, Turkey and South Africa offered weaker support as source populations for Jordan as evidenced by a lower F3 statistic (−0.0056).
The analysis by Whitfield et al. (2006) did not include either the Y group or A. cerana, though they used a composite root comprised of SNPs from A. cerana and A. dorsata. When we perform an nMDS analysis on a subset of the data that most closely reflects the set of populations in the Whitfield et al. (2006) analysis (fig. 3B and D ) we found that the A lineage is placed between A. cerana and the remainder of the A. mellifera populations. Moreover, the A lineage appears to be central to the M, C, and O lineages when A. cerana is not included, similar to figure 1A in Whitfield et al. (2006). This particular subset of the data would provide support for an African origin of A. mellifera, as concluded by Whitfield et al. (2006). Indeed, our analysis supports the idea that the European and Middle Eastern populations in the C, O, and M lineages are derived from the African populations, and that a substantial portion of diversification within A. mellifera occurred within Africa. However, a more comprehensive scenario emerges when additional populations are included.
We have provided multiple independent lines of evidence supporting the hypothesis that the origin of A. mellifera is located in Northeastern Africa or the Middle East with the ancestral A. mellifera population giving rise to the A and Y lineages, with further diversification and radiation of the C, O and M lineages from an African ancestral population. The precise placement of the A and Y lineages with respect to each other is not perfectly clear, in part because of lingering issues of data quality. In our ADMIXTURE analysis we see a clear differentiation between the Y population and the other A. mellifera populations. In addition, nMDS and FST analyses indicate differentiation between Y and the rest of A. mellifera. The TreeMix analyses, when comparing the entire data set to the subset of the data sequenced with Illumina, differ in the placement of the most basal node within A. mellifera. When all populations are considered, the Y lineage appears within the A lineage, whereas when only high quality sequence data is used, the Y and A lineages represent the most basal split within A. mellifera. We believe that these differences between trees are likely due to a higher residual error rate within the SOLiD sequenced individuals, and that the most basal node lies between the A and the Y lineages. Resequencing of O lineage and A. cerana individuals, the populations disproportionally affected by the low quality sequencing data, may resolve this placement. In addition, we suggest that further sampling of populations in northeastern Africa and the Middle East is necessary to better understand the origin of A. mellifera.
Formal tests of admixture can further reveal subtle relationships among populations. We find that many European populations show modest levels of admixture. The C group individuals from Austria and Germany and M group individuals from Norway and Sweden show low to moderate levels of admixture, which is unsurprising given the geographic proximity between the source populations and the admixed populations. These findings are consistently reconstructed by different methods including our TreeMix results, our ADMIXTURE results, and our ADMIXTOOLs analyses. These two lineages present interesting examples of continuing evolution in honey bees in Europe as these two distinct groups undergo gene flow. Their current geographic patterns may be driven by a number of factors, such as local adaptation or the influence of domesticated honey bees admixing with wild honey bees. The continued differentiation between the European C and M lineages and low levels of observed admixture, especially given the geographic proximity of these populations, makes them ideal candidates to further investigate patterns of local adaptation. The lack of enriched gene categories between these populations is also interesting as it suggests that there may be parallel patterns of evolution occurring in these lineages as they adapt to similar climates and other selective pressures within Europe.
More evidence for genic differences between populations can be seen in our analysis of the distributions of SNPs within the genome. We find many more SNPs in exonic regions than we would expect from a random distribution of SNPs throughout the genome, and we also detected hundreds of non-synonymous SNPs that could reflect functional differences in proteins. Our sets of highly differentiated SNPs between populations also revealed genes that have been studied extensively for their roles in honey bee biology such as vitellogenin and the major royal jelly proteins. We also find that there are several enriched gene ontology categories between multiple pairs of populations when we consider the 95th percentile of FST differences. Some of these terms, like transmembrane helix and sensory transduction, appear in a number of population pair comparisons suggesting that genes associated with these loci may frequently experience selective pressures as populations spread through various habitats. Sensory transduction refers to the translation of input stimuli to a signal received by the brain. Changes in these genes may result in adaptation to different food sources or other resources in novel environments. The term olfaction, which appears in three of our comparisons, may also be related to the detection of chemical stimuli associated with food resources. These sets of non-synonymous SNPs, high FST differences and gene ontology categories provide multiple sets of candidate genes that can be used in future research on local adaptation to climate and could be linked to honey bee health and breeding management.
We have presented evidence for a complex demographic history for A. mellifera that encompasses multiple instances of colonization and diversification. We propose a geographic origin of A. mellifera in the Middle East/Northeastern Africa with an ancestral population most closely related to the modern A and Y lineages. Subsequent radiations out of Africa into other parts of the Middle East and Europe gave rise to the other major lineages to produce regions of admixture in the Middle East where diverged population made secondary contact. This history is difficult to disentangle without a large, diverse data set capable of identifying differences between and relationships among many current populations.
This evolutionary history produced a series of population expansions into new regions with distinct climatic regimes. As honey bees colonized these new regions, numerous genomic regions must have experienced functional changes as honey bees experienced novel selective pressures. We have identified multiple genes that show differentiation between populations as well as gene ontology categories that differ between populations. These genomic regions may be used in future studies that aim to understand the genetic basis of adaptation to climate and could be used to improve managing practices.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the Berkeley Initiative for Global Change Biology, the Office of the Vice Chancellor for Research at UC Berkeley, the Gordon and Betty Moore Foundation (GBMF2983), the Vincent Coates Genome Sequencing Facility (NIH S10 Instrumentation Grants S10RR029668 and S10RR027303), and the USDA National Institute of Food and Agriculture, Hatch Project (CA-B-INS-0087-H).
Literature Cited
- Aizen MA, Garibaldi LA, Cunningham SA, Klien AM. 2009. How much does agriculture depend on pollinators? Lessons from long-term trends in crop production. Ann. Bot. 103:1579–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alburaki M, Moulin S, Legout H, Alburaki A, Garnery L. 2011. Mitochondrial structure of Eastern honeybee populations from Syria, Lebanon and Iraq. Apidologie. 42:628–641. [Google Scholar]
- Alburaki M, et al. 2013. A fifth major genetic group among honeybees revealed in Syria. BMC Genet. 14:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DJ, Novembre J, Lange K. 2009. Fast model-based estimation of ancestry in unrelated individuals. Gen. Res. 19:1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias MC, Sheppard WS. 2005. Phylogenetic relationships of honey bees (Hymenoptera:Apinae:Apini) inferred from nuclear and mitochondrial DNA sequence data. Mol. Phylo. Evol. 37:25–35. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. Ser. B. 57:289–300. [Google Scholar]
- Calderone NW. 2012. Insect pollinated crops, insect pollinators and US agriculture: trend analysis data for the period 1992-2009. PLoS One doi:10.1371/journal.pone.0037235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, et al. 2016. Genomic analyses reveal demographic history and temperate adaptation of the newly discovered honey bee subspecies Apis mellifera sinisxinyuan n. spp. Mol. Biol. Evol. doi:10.1093/molbev/msw017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapeau MD, Albert S, Kucharski R, Prusko C, Maleszka R. 2006. Evolution of the yellow/major royal jelly protein family and the emergence of social behavior in honey bees. Gen. Res. 16:1385–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels W, et al. 1990. Honeybee reproduction: Vitellogenin and caste-specific regulation of fertility In: Hoshi M, Yamashita O, editors. Advances in invertebrate reproduction 5. Amsterdam: Elsevier; p. 495–502. [Google Scholar]
- Franck P, Garnery L, Celebrano G, Solignac M, Cornuet JM. 2000. Hybrid origins of honeybees from Italy (Apis mellifera ligustica) and Sicily (A.m.sicula). Mol. Ecol. 9:907–921. [DOI] [PubMed] [Google Scholar]
- Franck P, et al. 2001. Genetic diversity of the honeybee in Africa: microsatellite and mitochondrial data. Heredity 86:420–430. [DOI] [PubMed] [Google Scholar]
- Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. 2009. Inferring the joint demographic history of multiple populations from multidimensional SNP data. PLoS Genet. 5:e1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F, Wallberg A, Webster MT. 2012. From where did the Western honeybee (Apis mellifera) originate? Ecol. Evol. 2:1949–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpur BA, et al. 2014. Population genomics of the honey bee reveals strong signatures of positive selection on worker traits. PNAS 7:2614–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009a. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 4:44–57. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009b. Bioinformatics enrichment tools: paths towards the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A-M, et al. 2007. Importance of pollinators in changing landscapes for world crops. Proc. R. Soc. B. 274:303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg S. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Meth. 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, 1000 Genome Project Data Processing Subgroup, et al. 2009. The sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, et al. 2012. Ancient admixture in human history. Genetics 192:1065–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Pritchard JK. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. e1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez SR, et al. 2010. A molecular phylogeny of the stingless bee genus Melipona (Hymenoptera: Apidae). Mol. Phyl. Evol. 56:519–525. [DOI] [PubMed] [Google Scholar]
- Rumble SM, et al. 2009. SHRiMP: accurate mapping of short color-space reads. PLoS Comput. Biol. 5(5):e1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruttner F. 1988. Biogeography and taxonomy of honeybees. New York: Springer-Verlag. [Google Scholar]
- Ruttner F, Tassencourt L, Louveaus J. 1978. Biometrical-statistical analysis of the geographic variability of Apis mellifera L. 1. Apidologie. 9:363–381. [Google Scholar]
- Sepylarskiy VB, Kharchenko P, Kondrashov AS, Bazykin GA. 2012. Heterogeneity of the transition/transversion ratio in Drosophila and Hominidae genomes. Mol. Biol. Evol. 29:1943–1955. [DOI] [PubMed] [Google Scholar]
- Sheppard WS. 1989. A history of the introduction of honey bee races into the United States, I and II. American Bee Journal 129:617–619–664–667. [Google Scholar]
- Sheppard WS, Mexiner MD. 2003. Apis mellifera pomonella, a new honey bee subspecies from Central Asia. Apidologie 34:367–375. [Google Scholar]
- South A. 2011. rworldmap: a new R package for mapping global data. R J. 3:35–43. [Google Scholar]
- vanEngelsdorp D, Meixner MD. 2010. A historical review of managed honey bee populations in Europe and the United States and the factors that may affect them. J. Inv. Path 103:580–595. [DOI] [PubMed] [Google Scholar]
- Wallberg A, et al. 2014. A worldwide survey of genome sequence variation provides insight into the evolutionary history of the honeybee Apis mellifera. Nat. Gen. doi:10.138/ng.3077 [DOI] [PubMed] [Google Scholar]
- Whitfield CW, et al. 2006. Thrice out of Africa: ancient and recent expansions of the honey bee Apis mellifera. Science 314:642–645. [DOI] [PubMed] [Google Scholar]
- Wilkin MB, et al. 2000. Drosophila Dumpy is a gigantic extracellular protein required to maintain tension at epidermal-cuticle attachment sites. Curr. Biol. 10:559–567. [DOI] [PubMed] [Google Scholar]
- Zheng X, et al. 2012. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinform. 28:3326–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.