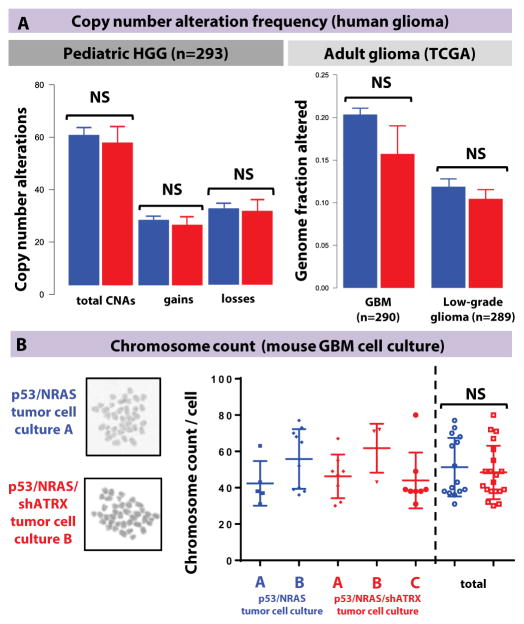
Fig. 4. ATRX loss does not cause structural/chromosomal alterations or change chromosome count.
(A) Analysis of chromosomal alterations, including copy number alterations, in the integrated pediatric GBM datasets, and percentage of genomic alterations in the adult TCGA dataset shows no difference by ATRX mutational status. Line represents mean ± SEM; NS = > 0.05 using unpaired Mann-Whitney test. (B) Chromosome count by metaphase preparation of independent GBM neurosphere cultures. Line represents mean ± SEM; NS = > 0.05 using unpaired Mann-Whitney test. Both groups had similar coefficients of variation [31.4% in p53/NRAS tumor cells (n=15) and 30.7% in p53/NRAS/shATRX tumor cells (n=20)].