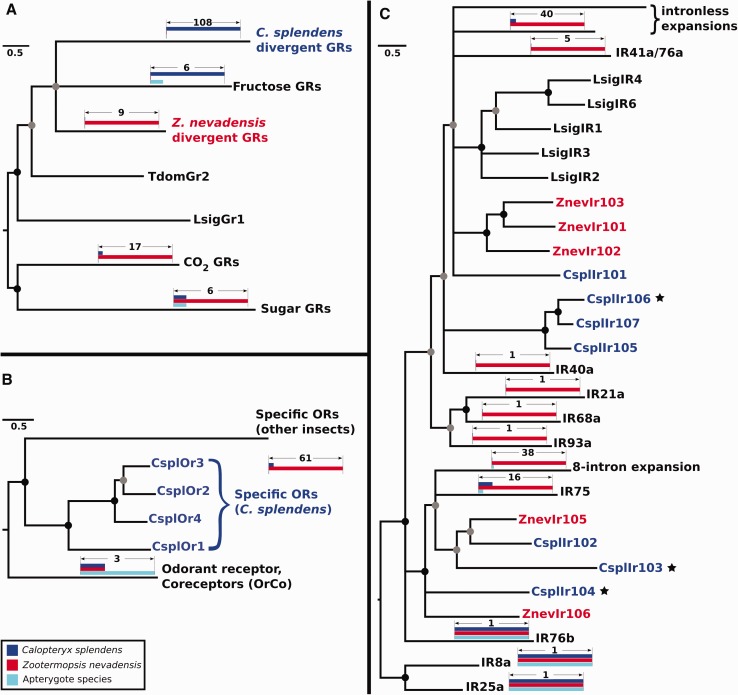
Fig. 4.—
Phylogenetic analysis of the chemoreceptors identified in the genome of Calopteryx splendens. (A) The majority of the 115 gustatory receptors (GRs) belong to a species-specific expansion whose specificity is unknown. There are also genes with similarity to GRs for recognizing fructose, CO2, and sugars; (B) C. splendens contains only five odorant receptors (ORs). One of them is the conserved OrCo protein, whereas the remaining four are specific ORs, which appear as a sister group to specific ORs from other insect species. (C) Many conserved ionotropic receptors (IRs) were identified in the damselfly genome, in addition to an equal number of divergent IRs. In all three panels, damselfly genes are colored in blue and genes from the termite Zootermopsis nevadensis are colored in red. Transcribed genes are indicated with a star next to them. Nodes with <50% bootstrap support collapsed into multifurcating nodes, nodes with bootstrap support between 50% and 75% are indicated with gray circles, and nodes with bootstrap support >75% are indicated with black circles. Branch length scale is in substitutions per site. Abbreviations used for species names: Cspl, C. splendens; Znev, Zootermopsis nevadensis; Tdom, Thermobia domestica; Lsig, Lepismachilis y-signata.