Figure 1. RAD51AP1-UAF1 complex formation via the SIM and SLD1-SLD2 domains in these proteins.
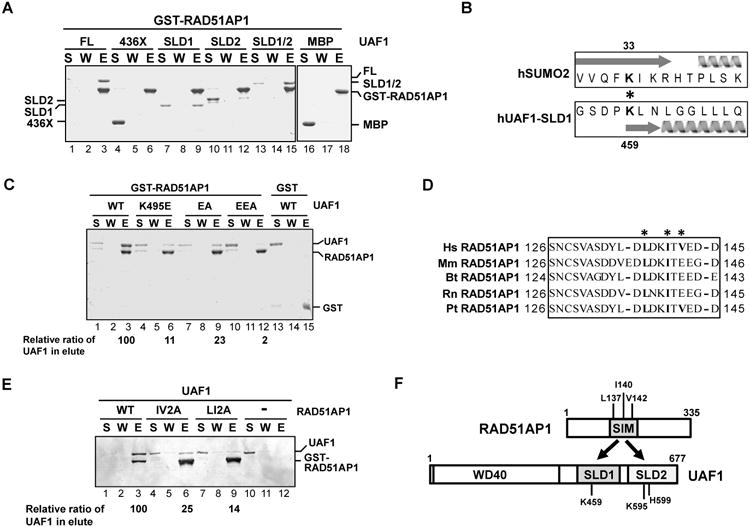
(A) Strep II-tagged UAF1, UAF1-436X, MBP-tagged UAF1-SLD1, UAF1-SLD2, and UAF1-SLD1/SLD2 (SLD1/2) were incubated with GST-tagged RAD51AP1, and protein complexes were captured on glutathione resin and analyzed by SDS-PAGE. S, supernatant containing unbound proteins; W, wash; E, SDS eluate of the glutathione resin.
(B) Alignment of UAF1-SLD1 against SUMO2 is shown. The asterisk highlights the K459 and K33 residues in UAF1 and SUMO2, respectively. Arrow and helix represent beta sheet and alpha helix, respectively.
(C) GST-tagged RAD51AP1 was incubated with Strep II-tagged UAF1 (WT) or the indicated UAF1 mutant, and protein complexes were captured with glutathione resin. Analysis was as in (A).
(D) Sequence analysis reveals a SIM between amino acid residues 137-142 in RAD51AP1. The asterisks highlight the residues targeted for mutagenesis. Hs, Homo sapiens; Mm, Mus musculus; Bt, Bos taurus; Rn, Rattus norvegicus; Pt, Pan troglodytes.
(E) GST-tagged RAD51AP1 (WT) or the indicated RAD51AP1 mutant was incubated with Strep II-tagged UAF1, and protein complexes were captured with glutathione resin. Analysis was as above.
(F) Schematic highlighting the RAD51AP1-UAF1 interaction domains.
See also Figure S1.
