Abstract
Buruli ulcer (BU) is an insidious neglected tropical disease. Cases are reported around the world but the rural regions of West and Central Africa are most affected. How BU is transmitted and spreads has remained a mystery, even though the causative agent, Mycobacterium ulcerans, has been known for more than 70 years. Here, using the tools of population genomics, we reconstruct the evolutionary history of M. ulcerans by comparing 165 isolates spanning 48 years and representing 11 endemic countries across Africa. The genetic diversity of African M. ulcerans was found to be restricted due to the bacterium’s slow substitution rate coupled with its relatively recent origin. We identified two specific M. ulcerans lineages within the African continent, and inferred that M. ulcerans lineage Mu_A1 existed in Africa for several hundreds of years, unlike lineage Mu_A2, which was introduced much more recently, approximately during the 19th century. Additionally, we observed that specific M. ulcerans epidemic Mu_A1 clones were introduced during the same time period in the three hydrological basins that were well covered in our panel. The estimated time span of the introduction events coincides with the Neo-imperialism period, during which time the European colonial powers divided the African continent among themselves. Using this temporal association, and in the absence of a known BU reservoir or—vector on the continent, we postulate that the so-called "Scramble for Africa" played a significant role in the spread of the disease across the continent.
Keywords: bacterial pathogen transmission, microbial population genomics, molecular evolution, phylogeography
Introduction
Buruli ulcer (BU) is a slowly progressing necrotizing disease of skin and subcutaneous tissue caused by the pathogen Mycobacterium ulcerans (Portaels et al. 2009). BU is considered a neglected tropical disease, and in some highly endemic areas, it is more prevalent than the most notorious mycobacterial diseases, tuberculosis, and leprosy (Walsh et al. 2011). Even though BU can affect all age groups, the majority of cases occur in children under age 15 (Debacker et al. 2004). On the African continent, the first detailed clinical descriptions of ulcers caused by M. ulcerans have been attributed to Sir Albert Cook, a missionary physician who worked in Uganda in 1897 (Cook 1970). Since this first description BU was reported in 16 Sub-Saharan African countries over the course of the 20th and the early 21th century. Today, more than 30 countries worldwide have reported the emerging disease, although the highest incidence by far is still observed in impoverished, rural communities of West and Central Africa (Janssens et al. 2005).
BU is known to occur primarily in foci around rural marshes, wetlands, and riverine areas (Wagner et al. 2008; Portaels et al. 2009). As proximity (but not contact) to these slow flowing or stagnant water bodies is a known risk factor for M. ulcerans infection (Jacobsen and Padgett 2010), it is generally believed that M. ulcerans is an environmental mycobacterium that can initiate infection after a micro-trauma of the skin (Meyers et al. 1974b; Williamson et al. 2014). Indeed, M. ulcerans DNA has been detected in a variety of aquatic specimens (Portaels et al. 1999; Vandelannoote et al. 2010), yet the significance of the detection of M. ulcerans DNA by PCR in environmental samples remains unclear in the disease ecology of BU (Portaels et al. 1999; Eddyani et al. 2004; Merritt et al. 2005; Marsollier et al. 2007; Williamson et al. 2008; Vandelannoote et al. 2010; Durnez et al. 2010; Gryseels et al. 2012). This is largely due to the fact that definite evidence for the presence of viable M. ulcerans in potential environmental reservoirs is lacking owing to the challenge of culturing the slow growing mycobacterium from non-clinical, environmental sources (Portaels et al. 2008). Consequently, the mode of transmission and non-human M. ulcerans reservoir(s) remain poorly understood (Röltgen and Pluschke 2015). As until today no animal reservoir for M. ulcerans has been identified in the Afrotropic ecozone (Durnez et al. 2010), our working hypothesis is that humans with active, openly discharging BU lesions may play a pivotal role in the spread of the bacterium.
Multilocus sequence typing analyses (Yip et al. 2007) and subsequent whole-genome comparisons (Doig et al. 2012) indicated that M. ulcerans evolved from a Mycobacterium marinum progenitor by acquisition of the virulence plasmid pMUM001. This plasmid harbors genes required for the synthesis of the macrocyclic polyketide toxin mycolactone (Stinear et al. 2004), which has cytotoxic and immunosuppressive properties that can cause chronic ulcerative skin lesions with limited inflammation and thus plays a key role in the pathogenesis of BU (George et al. 1999). Both the acquisition of the plasmid and a reductive evolution (Demangel et al. 2009; Stinear et al. 2007) suggested that a generalist proto-M. marinum became a specialized mycobacterium, more adapted to a restricted environment, perhaps within a vertebrate host. Analysis of the genome suggests that this new niche is likely to be protected from sunlight, non-anaerobic, osmotically stable, and an extracellular environment where slow growth, the loss of several immunogenic proteins, and production of the immunosuppressive molecule, mycolactone, provide selective advantages (Stinear et al. 2007; Doig et al. 2012). The evolution of M. ulcerans has been mediated by the insertion sequence element (ISE) IS2404, which is present in the M. ulcerans genome in ∼200 copies (Stinear et al. 2007). For some M. ulcerans lineages a second ISE, IS2606, is also present in a high copy number (∼90 copies). These short, mobile genetic DNA elements promote genetic rearrangements by modifying gene expression and sequestering genes (Mahillon and Chandler 1998). Increased ISE numbers are a signature for bacteria that have passed through an evolutionary bottleneck and undergone a lifestyle shift to a new niche, causing loss of genetic loci that are no longer required for the survival in the new environment (Moran and Plague 2004). Subsequent whole-genome comparisons showed that this "niche-adapted" genomic signature was established in a M. ulcerans progenitor before its intercontinental dispersal (Doig et al. 2012).
The restricted genetic diversity of M. ulcerans has meant that conventional genetic fingerprinting methods have largely failed to differentiate clinical disease isolates, complicating molecular analyses on the elucidation of the disease ecology, the population structure, and the evolutionary history of the pathogen (Roltgen et al. 2012). Whole genome sequencing (WGS) is currently replacing conventional genotyping methods for M. ulcerans (Doig et al. 2012; Ablordey et al. 2015; Bolz et al. 2015; Eddyani et al. 2015). Hence, in the present study, we sequenced and compared the genomes of 165 M. ulcerans disease isolates originating from multiple African disease foci to gain deeper insights into the population structure and evolutionary history of the pathogen, and to untangle the phylogeographic relationships within the genetically conserved cluster of African M. ulcerans.
Materials and Methods
Bacterial Isolates and Sequencing
We analyzed a panel of 165 M. ulcerans disease isolates originating from disease foci in 11 African countries that had been cultured between 1964 and 2012 (see supplementary table S1, Supplementary Material online). Isolates were chosen to maximize temporal and spatial diversity within countries in which more than 20 isolates were available (see supplementary fig. S1, Supplementary Material online). Even though most well-documented BU endemic countries were well represented, we were unable to include isolates from several African countries (Equatorial Guinea, Kenya, Liberia, Sierra Leone, and South Sudan), that have reported, if not isolated, (a limited number of) BU cases in the past (Janssens et al. 2005). Two isolates from Papua New Guinea (PNG) were included as out-groups to root the African phylogenetic tree. PNG M. ulcerans was specifically chosen as it (together with African and other Southeast Asian M. ulcerans) belongs to the more virulent and distinct "classic" phylogenetic lineage (Kaser et al. 2007), relative to M. ulcerans isolates elsewhere.
Permission for the study was obtained from the ITM Institutional Review Board. Isolates were processed and analyzed without use of any patient identifiers, except for country and village of origin if this information was available. Based on conventional phenotypic and genotypic methods, bacterial isolates had previously been assigned to the species M. ulcerans (WHO 2014). Mycobacterial isolates were maintained for prolonged storage at ≤−70 °C in Dubos broth enriched with growth supplement and glycerol. DNA was obtained by harvesting the growth of three Löwenstein–Jensen (LJ) slants followed by heat inactivation, mechanical disruption, enzymatic digestion, and DNA purification on a Maxwell 16 automated platform (Eddyani et al. 2015).
Index-tagged paired-end sequencing-ready libraries were prepared from gDNA extracts with the Nextera XT DNA Library Preparation Kit. Genome sequencing was performed on Illumina HiSeq 2000 and Miseq DNA sequencers according to the protocols of the manufacturers with 150bp, 250bp, or 300bp paired-end sequencing chemistry. Sequencing statistics are provided in supplementary table S1, Supplementary Material online. The quality of raw Illumina reads was investigated with FastQC v0.11.3 (Andrews 2015).
Prior to further analysis, reads were cleaned with clip, a tool in the Python utility toolset Nesoni v0.130 (Harrison and Seeman 2014). Reads were filtered to remove those containing ambiguous base calls, any reads <50 nucleotides in length, and reads containing only homopolymers. All reads were further trimmed to remove residual ligated Nextera adaptors and low quality bases (<Q10) at the 3' end. The total amount of read-pairs kept after clipping and their average read length are summarized for all isolates in supplementary table S1, Supplementary Material online.
Read Mapping and SNP/Large Deletion Detection
Read mapping and SNP detection were performed using the Snippy v2.6 pipeline (Seemann 2015). The Burrows–Wheeler Aligner (BWA) v0.7.12 (Li and Durbin 2009) was used with default parameters to map clipped read-pairs to two M. ulcerans Agy99 reference genomes: the M. ulcerans Agy99 bacterial chromosome (Genbank: CP000325) and the M. ulcerans pMUM001 plasmid (Genbank: BX649209). Due to the unreliability of read mapping in mobile repetitive regions all ISE elements (IS2404 and IS2606) and all (plasmid-encoded) polyketide synthase genes were masked in these reference genomes (397 kb/5.63 Mb—7% of Agy99, 118 kb/174 kb—67% of pMUM001). After read mapping to M. ulcerans Agy99 and pMUM001, average read depths were determined with SAMtools v1.2 (Li et al. 2009) and are summarized for all isolates in supplementary table S1, Supplementary Material online). SNPs were subsequently identified using the variant caller FreeBayes v0.9.21 (Garrison and Marth 2012), with a minimum depth of 10 and a minimum variant allele proportion of 0.9. Snippy was used to pool all identified SNP positions called in at least one isolate and interrogate all isolates of the panel at that position. As such a multiple sequence alignment of "core SNPs" was generated.
The number of reads mapping to unique regions of the plasmid and the bacterial chromosome was used to roughly infer plasmid copy number. This was achieved by calculating the ratio of the mode of read depth of all positions in the plasmid to that of the chromosome: Mo(read depth unique positions plasmid):Mo(read depth unique positions chromosome) (Holt et al. 2012).
Large (>1000 bp) chromosomal deletions were detected with Breseq v.0.27.1 (Barrick et al. 2014), a reference-based alignment pipeline that has been specifically optimized for microbial genomes. Breseq was used with Bowtie2 v.2.2.6 (Langmead and Salzberg 2012) to map clipped read-pairs to Agy99 and the resulting missing coverage evidence was used to detect large deleted chromosomal regions.
Population Genetic Analysis
Bayesian model-based inference of the genetic population structure was performed using the "Clustering with linked loci" module (Corander and Tang 2007) in BAPS v.6.0 (Corander et al. 2008). This particular module takes potential linkage within the employed molecular information into consideration, which is advisable when performing genetic mixture analysis on SNP data from a haploid organism. A concatenation of the core-SNP alignment of both the bacterial chromosome and the plasmid was loaded as a sequential BAPS formatted file. This entry was complemented with a "linkage map" file that differentiated two linkage groups (bacterial chromosome & plasmid). The optimal number of genetically diverged BAPS-clusters (K) was estimated in our data by running the estimation algorithm with the prior upper bound of K varying in the range of 3–20. Because the algorithm is stochastic, the analysis was run in 20 replicates for each value of K as to increase the probability of finding the posterior optimal clustering with that specific value of K.
QGIS v.2.10 (Quantum_GIS 2012) was used to generate the figures on the geographical distribution of African M. ulcerans. The residence of BU patients at the time of their clinical visit was represented as points. In the case where residence information was missing, we used the location of the hospital supplying the sample. The administrative borders of countries were obtained from the Global Administrative Unit Layers dataset of FAO.
Detection of Recombination
Evidence for recombination between different BAPS-clusters was assessed using several methods as studies show that no single method is optimal, whereas multiple approaches may maximize the chances of detecting recombination events (Posada and Crandall 2001). A whole genome alignment was constructed with Snippy using default parameters. Recombination was assessed using the pairwise homoplasy index test Φw (Bruen et al. 2006) (with significance set at 0.05), as implemented in Splitstree v 4.13.1 (Huson and Bryant 2006) on the whole genome alignment. We used BRAT-NextGen (Marttinen et al. 2012) to detect recombination events within our isolate panel using the whole genome alignment. BRAT-NextGen was specifically developed to detect homologous recombinant segments among a group of closely related bacteria over the process of their diversification and has been shown to be highly accurate when applied to mycobacteria (Mortimer and Pepperell 2014). The estimation of recombination was started with a partitioning of the whole genome alignment into 5kb segments and running a clustering analysis separately on each of these segments. The alpha hyper-parameter was estimated with default settings. The proportion of shared ancestry (PSA) tree was cut at 0.15 differentiating a total set of four clusters for all taxa. Recombination profiles were calculated with 100 iterations, at which stage parameter estimations had successfully converged. Significance (P < 0.05) of each putative recombinant segment was determined with 100 pseudo replicate permutations.
Maximum-Likelihood Phylogenetic Analysis
A maximum-likelihood (ML) phylogeny was estimated 10 times from the SNP alignment using RAxML v8.2.0 (Stamatakis 2014) under a plain generalized time reversible (GTR) model (no rate heterogeneity) with likelihood calculation correction for ascertainment bias using the Stamatakis method (Stamatakis 2015). Identical sequences were removed before the RAxML runs. For each run, we performed 10,000 rapid bootstrap analyses to assess support for the ML phylogeny. The tree with the highest likelihood across the 10 runs was selected. We used TreeCollapseCL v4 (Hodcroft 2013) to collapse nodes in the tree with bootstrap values below a set threshold of 70% (Hillis and Bull 1993) to polytomies, whereas preserving the length of the tree. Phylogenetic relationships were inferred with the two PNG strains as outgroups. Root-to-tip distances were extracted from the ML phylogeny using TreeStat v1.2 (Rambaut 2008). The relationship between root-to-tip distances and tip dates (Rieux and Balloux 2016) was determined using linear regression analysis in R v3.2.0 (R Core Team 2015).
Bayesian Phylogenetic Analysis
We used BEAST2 v2.2.1 (Bouckaert et al. 2014) to date evolutionary events, determine the substitution rate and produce a time-tree of African M. ulcerans, as this approach allows for inference of phylogenies with a diverse set of molecular clock and population parameters (Drummond et al. 2006). Path sampling (PS) (Lartillot and Philippe 2006) was used to determine the best clock and population model priors by computing the marginal likelihoods of competing models of evolution, as this method has been shown to outperform other methods of model selection (Baele et al. 2012; Lartillot and Philippe 2006). We compared three clock models (strict, uncorrelated exponential relaxed, and uncorrelated log-normal relaxed) in combination with two demographic coalescent models (constant and exponential). The required number of steps in PS analysis was determined by running one of the more complex models (uncorrelated log-normal relaxed clock/constant coalescent tree prior) with a different amount of steps, starting from 100 steps until 400 using increments of 50. As no difference in marginal likelihood estimates was observed after 100 steps, each model was run for 100 path steps, each with 200 million generations, sampling every 20,000 MCMC generations and with a burn-in of 30%. Likelihood log files of all individual steps were inspected with Tracer v1.6 (Rambaut et al. 2014) to see whether the chain length produced an effective sample size (ESS) larger than 400, indicating sufficient sampling. Marginal likelihoods of the models were then used to calculate natural log Bayes factors (LBF = ln mL(model1)−mL(model2)), which evaluate the relative merits of competing models (see also supplementary material, Supplementary Material online).
The best clock/demographic model (UCLD relaxed clock with a constant coalescent tree prior—see also supplementary material, Supplementary Material online) was then used to infer a genome scale African M. ulcerans time-tree under the GTR substitution model and with tip-dates defined as the year of isolation (supplementary table S1, Supplementary Material online). Analysis was performed in BEAST2 using a total of 10 independent chains of 200 million generations, with samples taken every 20,000 MCMC generations. Log files were inspected for convergence and mixing with Tracer v1.6. LogCombiner v2.2.1 (Bouckaert et al. 2014) was then used to combine log and tree files of the independent BEAST2 runs, after having removed a 30% burn-in from each run. Thus, parameter medians and 95% highest posterior density (HPD) intervals were estimated from over 1.6 billion visited MCMC generations. To ensure prior parameters were not over-constraining the calculations, the entire analysis was furthermore run while sampling only from the prior. Finally, we also checked for the robustness of our findings under different priors, as states of low mutation rate and large troot are hard to distinguish from otherwise identical states of large mutation rate and smaller troot (Drummond et al. 2002). This additional analysis was required as the tree prior and the clock prior interact when adding sequence data, and the strength of this interaction is not visible when sampling exclusively from the prior.
TreeAnnotator was used to summarize the posterior sample of time-trees so as to produce a maximum clade credibility tree with the posterior estimates of node heights visualized on it (posterior probability limit ≥ 0.8).
A permutation test was used to assess the validity of the temporal signal in the data. This was undertaken by performing 20 additional BEAST2 runs (of 200 million MCMC generations each) with identical substitution (GTR), clock (uncorrelated log-normal relaxed), and demographic models (constant coalescent) but with tip dates randomly reassigned to sequences. This random "null set" of tip-date and sequence correlations was then compared with the substitution rate estimate of the genuine tip-date and sequence correlations (Holt et al. 2012; Rieux and Balloux 2016).
Discrete Phylogeographic Analysis
To assess the geospatial distribution of African M. ulcerans through time, an additional BEAST2 analysis was performed. In this analysis, the posterior probability distribution of the location state (geographic region) of each node in the tree was inferred, in addition to the parameters described above (tree topology, evolutionary, and demographic model). Sampled isolates were associated with fixed discrete location states and a discrete BSSVS geospatial model (Lemey et al. 2009) was subsequently used to reconstruct the ancestral location states of internal nodes in the tree from these isolate regions. To prevent loss of the signal in the data by considering too many discrete regions compared with the number of isolates, we limited the amount of discrete regions by merging neighboring countries (similarly as carried out in Comas et al. 2013; He et al. 2013). As such we differentiated five regions: Ivory Coast (20 isolates), Ghana-Togo (25 isolates), Benin-Nigeria (65 isolates), Cameroon-Gabon (24 isolates), and Angola-DRCongo-Congo (30 isolates). Ten independent chains were run for 200 million generations, with subsamples recorded from the posterior every 20,000 MCMC generations. LogCombiner was then used to combine tree files of the independent BEAST2 runs, after having removed a 30% burn-in from each run. TreeAnnotator was used as described above to summarize the posterior sample of time-trees.
Results and Discussion
In order to understand how and when M. ulcerans has spread across Africa, we sequenced the genomes of 165 African isolates that were obtained between 1964 and 2012 and spanned most of the known endemic areas of BU in 11 different African countries (supplementary fig. S1, Supplementary Material online). This collection captured as much diversity as possible within Africa whereas minimizing the phylogenetic discovery bias implicit to SNP typing (Pearson et al. 2004; Roltgen et al. 2010). Resulting sequence reads were mapped to the Ghanaian M. ulcerans Agy99 reference genome and, after excluding mobile repetitive IS elements and small insertion-deletions (indels), we detected a total of 9,193 SNPs uniformly distributed across the M. ulcerans chromosome with approximately 1 SNP per 613 bp (0.17% nucleotide divergence) (supplementary fig. S2, Supplementary Material online). Similarly, a total of 81 SNPs were identified in the non-repetitive regions of pMUM001, which resulted in a very comparable nucleotide divergence of 0.14%. The maximum chromosomal genetic distance between two African isolates was 5,157 SNPs.
The Population Structure of M. ulcerans in Africa
Large DNA deletions are excellent evolutionary markers because they are very unlikely to occur independently in different lineages but rather are the result of unique irreversible events in a common progenitor (Brosch et al. 2002). We explored the distribution of large chromosomal deletions (relative to Agy99) and identified differential genome reduction that supports the existence of two specific M. ulcerans lineages within the African continent, hereafter referred to as Lineage Africa I (Mu_A1) and Lineage Africa II (Mu_A2). A total of 20 genomic deletion events were identified in all lineage Mu_A2 strains (table 1). Most of the 20 deletions were mediated by IS elements IS2404 and IS2606 which are known to profoundly enhance mycobacterial genome plasticity (Stinear et al. 2007). These high copy number IS elements either flanked the deletion or they were present in the deleted or substituted sequence stretches. The 20 identified deletions contained a total of 27 protein coding sequences (CDSs) and 12 pseudogenes (table 1). The number of deleted CDSs and pseudogenes averaged two per deletion event (range one–eight). Well represented were pseudogenes that either contained frameshift mutations or were disrupted by IS elements.
Table 1.
Genomic Deletions in Mu_A2 Relative to the Agy99 Reference Genome (Mu_A1).a in Agy99
| Start positiona | End positiona | Size of deletion (bp) | Description of affected genesa |
|---|---|---|---|
| 26779 | 28225 | 1446 | MUL_0025: transposase for IS2606 |
| 132163 | 133609 | 1446 | MUL_0131: transposase for IS2606 |
| 826566 | 828021 | 1455 | MUL_0780: transposase for IS2606 |
| 1618246 | 1619692 | 1446 | MUL_1494: hypothetical cytoplasmic protein; MUL_1495: transposase for IS2606 |
| 1711639 | 1713085 | 1446 | MUL_1581: conserved hypothetical protein (pseudogene—disrupted by IS2404); MUL_1582: transposase for IS2404 |
| 2627839 | 2629851 | 2012 | MUL_2348: polyketide synthase (pseudogene—disrupted by IS2606 & Frame shift mutation) |
| 2886326 | 2887702 | 1376 | MUL_2586: PPE family protein (pseudogene—disrupted by IS2404); MUL_2585: transposase for IS2404 |
| 2976946 | 2980459 | 3513 | MUL_2663: metal cation transporting p-type ATPase CtpH_1 (pseudogene—Frame shift mutation) |
| 3055344 | 3058692 | 3348 | MUL_2733: conserved hypothetical protein (pseudogene—disrupted by IS2404); MUL_2735: transposase for IS2606; MUL_2736: transposase for IS2404 |
| 3304274 | 3307178 | 2904 | MUL_2983: transposase for IS2606; MUL_2984: transposase for IS2404 |
| 3594994 | 3596440 | 1446 | MUL_3219: type I restriction-modification system restriction subunit (pseudogene—frame shift mutation); MUL_3221: transposase for IS2606 |
| 3603880 | 3605326 | 1446 | MUL_3229: transposase for IS2606 |
| 3811852 | 3813298 | 1446 | MUL_3438: transposase for IS2606 |
| 3986318 | 3989256 | 2938 | MUL_3592: transposase for IS2606 (pseudogene—frame shift mutation); MUL_3593: transposase for IS2404 |
| 4286753 | 4288200 | 1447 | MUL_3836: transposase for IS2606 (pseudogene—frame shift mutation) |
| 4687633 | 4689079 | 1446 | MUL_4214: transposase for IS2606; MUL_4215: N-term cell filamentation protein Fic (pseudogene—disrupted by insertion sequence) |
| 4848769 | 4850215 | 1446 | MUL_4368: transposase for IS2606 |
| 4876318 | 4879973 | 3655 | MUL_4394: hypothetical membrane protein; MUL_4395: mid-section conserved hypothetical membrane protein (pseudogene—DNA deletion); MUL_4397: transposase for IS2606; MUL_4398: transposase for IS2404 |
| 4961244 | 4962690 | 1446 | MUL_4474: transposase for IS2606 |
| 5088793 | 5096926 | 8133 | MUL_4588: N-term conserved hypothetical protein (pseudogene—disrupted by IS2404); MUL_4589: transposase for IS2404; MUL_4590: hypothetical cytoplasmic protein; MUL_4591: hypothetical cytoplasmic protein; MUL_4592: hypothetical cytoplasmic protein; MUL_4593: conserved hypothetical protein (pseudogene—frame shift mutation); MUL_4594: hypothetical cytoplasmic protein; MUL_4595: transposase for IS2404 |
SNP-based exploration of the genetic population structure using BAPS (Corander et al. 2008) agreed with the above deletion analysis and subdivided the African M. ulcerans population into four major clusters. Clusters 1–3 constitute Mu_A1 whereas BAPS-cluster 4 corresponds to Mu_A2. The composition of these clusters is detailed in supplementary table S1, Supplementary Material online. Cluster 1 circulates throughout the African continent and represents the vast majority of the isolates, n = 136 (82.4%). This cluster is also the most genetically diverse with an intra-cluster average pairwise SNP difference (SNPΔ) of 171 SNPs (SD = 73). Clusters 2, 3, and 4 were considerably smaller, encompassing 20 (12.1%), 1 (0.6%), and 8 (4.8%) isolates, respectively. Cluster 2 circulates in different regions of Cameroon and neighboring Gabon and corresponds to an SNPΔ of 64 SNPs (SD = 54). Cluster 3 (1 isolate) was only found in Uganda. Finally, Cluster 4 (which encompasses Mu_A2 entirely) has a SNPΔ of 81 SNPs (SD = 41). Relative to the occurrence of Mu_A1, Mu_A2 is common in Gabon (40%, 2 Mu_A2/5 total Gabonese strains), but quite rare in Cameroon (5%, 1/19) and Benin (8%, 5/59).
African M. ulcerans Evolves through Clonal Expansion, Not Recombination
Ignoring recombination when analyzing evolving bacterial pathogens can be misleading as the process has the potential to both distort phylogenetic inference and create a false signal of apparent mutational evolution by (horizontally) introducing additional divergence between heterochronously sampled disease isolates (Croucher et al. 2013). The pairwise homoplasy index (Φw) did not find statistically significant evidence for recombination (P= 0.1545) between different BAPS-clusters. Correspondingly, BRAT-NextGen did not detect any recombined segments in any isolate, supporting a strongly clonal population structure for M. ulcerans that is evolving by vertically inherited mutations.
Phylogenetic Analysis Reveals Strong Geographical Restrictions on M. ulcerans Dispersal
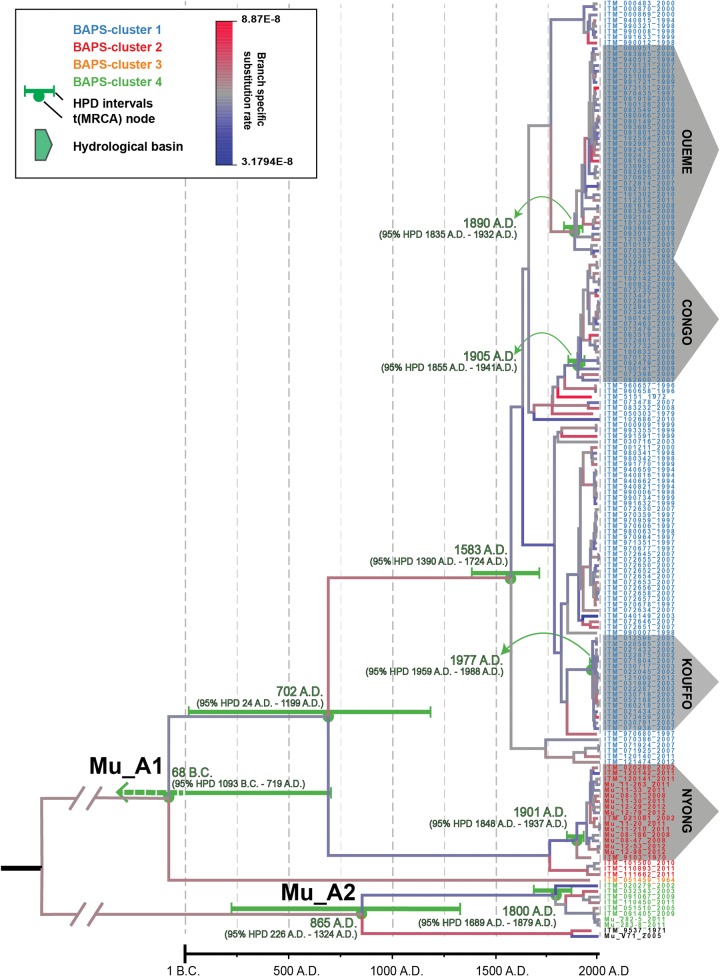
A phylogeny was reconstructed from the chromosomal SNP alignment using both maximum-likelihood (RAxML) and Bayesian (BEAST2) approaches. Supplementary fig. S3, Supplementary Material online shows a well-supported ML-phylogeny that resolved the two major African lineages Mu_A1 and Mu_A2 and distinguished between the four BAPS-clusters within the African panel. Both the lineages and the BAPS-clusters had 100% bootstrap support and a Bayesian posterior probability of 1 (BEAST2 tree—fig. 1). The genome-based phylogeny was consistent with previously constructed phylogenies based on discriminating ISE-SNP markers even though these previous trees suffered from low branch support (Vandelannoote et al. 2014). The tree also indicated that Mu_A1 is much more widely dispersed within the African continent than Mu_A2.
Fig. 1.—
Bayesian maximum clade credibility phylogeny for African M. ulcerans. The tree was visualized and colored in Figtree v1.4.2 (Rambaut 2015). Branches are color coded according to their branch specific substitution rate (legend at top). Branches defining major lineages are annotated on the tree. Tip labels are color coded according to their respective BAPS-clusters (the best visited BAPS partitioning scheme of our sample yielded a natural log marginal likelihood of − 95,857). Divergence dates (mean estimates and their respective 95% HDP) are indicated in green for major nodes. Note 95% HDP intervals grow larger closer to the root of the tree as increasingly less timing calibration information is available the further one goes back in time. Geographically localized clonal expansions associated with four particular hydrological basins (Congo, Kouffo, Oueme, and Nyong) are highlighted with boxes and their corresponding t(MRCA) & 95% HDP are specified in green.
We identified an unambiguous relationship between the genotype of an isolate and its geographical origin. This is illustrated by the explicit regional clustering of M. ulcerans within the phylogenetic tree, indicating significant geographical structure in the African mycobacterial population. For instance, all strains isolated from patients living in the hydrological basin of the Kouffo River of southern Benin clustered together in a "basin-specific" clade in the Bayesian phylogeny (fig. 1). Our observations confirm and extend previous data showing geographical subdivisions (Doig et al. 2012; Roltgen et al. 2010; Vandelannoote et al. 2014; Ablordey et al. 2015; Bolz et al. 2015), and indicate that when M. ulcerans is introduced in a particular area, it remains isolated and localized for a sufficient amount of time to allow mutations to become fixed in that population. As such, a local genotype that is associated with that area is allowed to evolve.
We also identified a strong association between the distribution of particular genotypes and hydrological drainage areas. It appears that the borders of hydrological basins (consisting of elevated regions, and salt water) also form a barrier to bacterial spread. For instance, the isolates of the Kouffo Basin are distinct from isolates originating from the neighboring Oueme Basin, and are in fact more related to isolates from Ghana and Ivory Coast (fig. 1).
A Central Role for the Mycolactone Producing Plasmid
All sequenced isolates carried the pMUM001 plasmid. By comparing read depths of all plasmid and chromosome positions we roughly estimated an average pMUM001 copy number of 1.3 copies per cell (range 0.4–1.7). Furthermore, the plasmid ML-tree (built with the discovered 81 SNPs) was observed to closely match the topology and relative branch lengths of the chromosome ML-tree (supplementary fig. S3A, Supplementary Material online), which is consistent with co-evolution of the plasmid with the host-chromosome, stable maintenance of the plasmid, and absence of transfer of plasmid variants between host bacteria. This asserted the central role of the mycolactone producing plasmid in the evolution of African M. ulcerans. These findings, combined with the observation that mycolactone negative isogenic mutants are avirulent (George et al. 1999), indicate that there is strong purifying selection on maintaining a functional plasmid that allows the mycobacterium to biosynthesize its primary virulence determinant mycolactone.
The Substitution Rate of African M. Ulcerans Is Remarkably Low
The major objective of this study was to estimate the rate of evolution of M. ulcerans in order to estimate the temporal dynamics of the spread of the pathogen across Africa. Like Mycobacterium tuberculosis (Comas et al. 2013), M. ulcerans does not exhibit a strict molecular clock with substitution mutations occurring at a fixed regular rate, complicating temporal inferences. To overcome this potential limitation, we used a Bayesian approach with a relaxed molecular clock model to infer the evolutionary dynamics of the African mycobacterial population (see also supplementary material, Supplementary Material online). As such, a genome scale African M. ulcerans time-tree was inferred (fig. 1), whereas also providing estimates of nucleotide substitution rates and divergence times for key M. ulcerans clades.
We estimated a mean genome wide substitution rate of 6.32E−8 per site per year (95% HPD interval [3.90E−8 to 8.84E−8]), corresponding to the accumulation of 0.33 SNPs per chromosome per year (95% HPD interval [0.20−0.46]) (excluding IS elements). To test the validity of the discovered temporal signal in the data, we performed 20 permutation tests. This produced a null set of 20 "randomized" substitution rate distributions, which were significantly different (Wilcoxon test, P < 2.2E−16) to the substitution rate estimate of the genuine tip-date and sequence correlation (see supplementary fig. S4, Supplementary Material online). This clearly indicated that the tip dates were informative and could provide sufficient calibrating information for the analysis (Holt et al. 2012; Rieux and Balloux 2016). The estimated genome wide substitution rate is lower than the estimate for Clostridium difficile (1.88E−7) (He et al. 2013) and Shigella sonnei (6.0E−7) (Holt et al. 2012) yet slightly higher than that of M. tuberculosis (2.6E−9), the bacterium with the slowest rate currently described (Comas et al. 2013). The analysis also indicated that the genealogy has undergone very moderate rate variation, with a 2.8-fold difference between the slowest (3.18E−8) and the fastest (8.87E−8) evolving branches. Rate accelerations and decelerations are found interspersed in the time-tree (fig. 1). The observed slight rate variation is probably attributable to fluctuations in the number of bacterial replication cycles per time unit, changes in selection pressures through time, or combinations of these factors.
M. ulcerans Has Existed in Africa for Centuries and was Recently Re-Introduced
African Mu_A2 strains were found to form a very strongly supported (posterior probability = 1) monophyletic group with two PNG strains that were included in the analysis as an outgroup, indicating a closer relationship with strains from PNG than African Mu_A1 stains. The Bayesian analysis (fig. 1) indicated furthermore that lineage Mu_A1 has been endemic in the African continent for hundreds of years (tMCRA(Mu_A1) = 68 BC (95% HPD 1093 BC–719 AD)). Conversely, Lineage Mu_A2 was derived to have been introduced much more recently in the African continent (tMCRA(Mu_A2) = 1800 AD (95% HPD 1689 AD–1879 AD)), explaining why the lineage is less common and more geographically restricted. Interestingly, the estimated time span of the Mu_A2 introduction event coincides with a historical event of particular interest: the period of Neo-imperialism (late 19th–early 20th century). During this period, the European powers divided the African continent among themselves through the invasion, colonization, and annexation of territory. In the absence of a known alternative reservoir, nor vector, this specific temporal association implies a human-mediated Mu_A2 introduction event, whether through the introduction of M. ulcerans bacteria within diseased humans, or an alternative reservoir or vector.
Recent Introduction of M. ulcerans in the Congo, Kouffo, Oueme, and Nyong Basins
The time-tree of African M. ulcerans also reveals evidence of the potential role that the so-called "Scramble for Africa" played in the spread of endemic Mu_A1 clones in three hydrological basins (Congo, Oueme, and Nyong) that are particularly well covered by our isolate panel (fig. 1) (Pakenham 1993). Because, to our knowledge, no epidemiological studies were conducted in these hydrological basins until the late 1900s, whether BU was a newly introduced, versus an old expanding illness in these regions (Janssens et al. 2005) had remained unclear to date. Close inspection of the time-tree implied that, similar to the Mu_A2 introduction event, the basin-specific Mu_A1 introduction events coincide with the start of colonial rule (table 2).
Table 2.
Timing of Introduction Events of Four Selected Epidemic Lineage Mu_A1 Clones in Their Respective Hydrological Basin
| Hydrological basin | Endemic hotspot | Approx. start of colonial rule | Mean t(MRCA) | 95% HPD t(MRCA) | First reported cases |
|---|---|---|---|---|---|
| Congo | Songololo Territory | 1885a | 1905 | 1855–1941 | 1961d (Andersen 1965) |
| Nyong | Between Ayos and Akonolinga | 1884c | 1901 | 1848–1937 | 1969 (Ravisse 1977; Ravisse et al. 1975) |
| Oueme | Southeastern Benin | 1892b | 1890 | 1835–1932 | 1988 (Muelder and Nourou 1990) |
| Kouffo | Southeastern Benin | 1892b | 1977 | 1959–1988 | 1977 (Aguiar 1997) |
Founding of the Belgian Congo Free State.
Kingdom of Dahomey annexed into the French colonial empire.
German Empire claimed the colony of Kamerun and began a steady push inland.
Based on interviews and observations of healed lesions in the villages of the Songololo territory it was believed that M. ulcerans infections already existed in the area in 1935 (Meyers et al. 1974a). HDP, highest probability density interval; t(MRCA), time to most recent common ancestor.
To situate the historical model we are suggesting here, it is important to note that inhabitants of the three regions of interest have long exploited the river and forest ecologies prior to the arrival of the European colonial powers (Giles-Vernick et al. 2015). Many of the basin’s inhabitants relied on natural resources for their survival and as such, were continuously exposed to the lentic environments. However, it was only after the start of colonial rule that the basin associated epidemic Mu_A1 clones were introduced, presumably through the introduction of a M. ulcerans reservoir or vector. However, given the fact that no vector or reservoir species is known in the Afrotropic ecozone other than Homo sapiens (Durnez et al. 2009; Vandelannoote et al. 2010; Narh et al. 2015), we postulate that it was the arrival of displaced BU-infected humans that played a role in the observed spread of M. ulcerans. Colonialism was commonly violent and introduced significant socio-economic changes in the three basins that often involved population displacement. In all likelihood, displaced BU-infected humans were not directly infecting other humans as human-to-human transmission of M. ulcerans is extremely rare (Debacker et al. 2003). Humans were nevertheless in all probability an important reservoir as displaced BU-infected patients with active, openly discharging lesions could contaminate the environment during water contact activities by shedding concentrated clumps of mycobacteria.
A fourth noteworthy hydrological basin is that of the Beninese Kouffo River. The timing of its basin specific introduction event (1977 AD (95% HPD 1959 AD–1988 AD)) is much more recent than the three previously discussed basins. Notably, the first BU cases from this region were identified and treated in 1977 (Aguiar 1997), concurrent with the estimated introduction event.
The Historic Spread of M. ulcerans Lineage Mu_A1
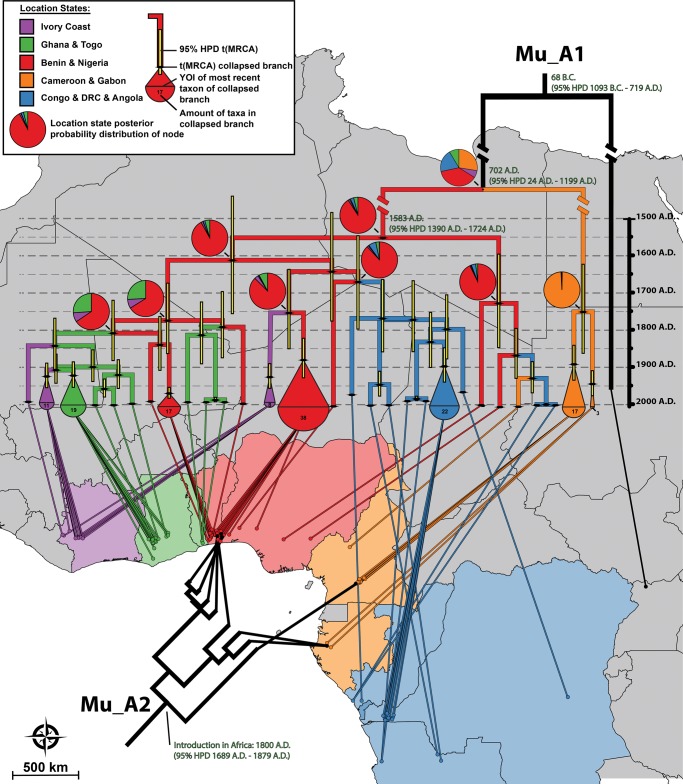
The phylogeographic analysis also offered new insights on the geospatial spread of M. ulcerans lineage Mu_A1 through time (fig. 2). Uganda, Cameroon, and Gabon occupy basal branches in the Mu_A1 time-tree indicating that the bacterium has been extant in these regions for the longest time. The lineage subsequently expanded from these regions into West and Central Africa. Ancestral state reconstruction analysis indicated that this expansion most likely occurred from the region that encompasses Benin and Nigeria (posterior probability = 0.91). From there, Mu_A1 spreads further west (into Togo, Ghana, and Ivory Coast) and back east (into Congo, DRC, and Angola).
Fig. 2.—
Geospatial distribution of African M. ulcerans through time. A Bayesian maximum clade credibility phylogeny is drawn for lineage Mu_A1 with branches color coded according to their most likely location state (legend at top). Pie charts indicate location state posterior probability distributions of major nodes. The amount of location states was limited to five by merging the disease isolates of certain neighboring countries. The genetically distinct Ugandan singleton node (which represents its own BAPS-cluster) was omitted from the analysis as multiple isolates are required per cluster. Divergence dates (mean estimates and their respective 95% HDP) are indicated in green for nodes that fall outside of the time scale. A number of oversampled localized clonal expansions are collapsed in the tree with the size of their representing cartoon proportional to the amount of collapsed taxa. The tips of the tree are connected to the location of residence of patients from whom the isolate was grown.
In other bacterial pathogens, the discipline of microbial phylogeography has also proved to be a powerful means of investigating not only the spread of microbes but also the movement of their hosts. For example, interesting associations were found between the genotypes of Helicobacter pylori strains, their places of origin, and the migration and ethnicity of their human hosts (Linz et al. 2007). Additionally, the spread of M. tuberculosis and M. leprae also reflects the migrations of early humans (Monot et al. 2009; Comas et al. 2013).
Comparable as in these landmark studies, the phylogeographic approach applied here is limited in the respect that it studies a sample of the mycobacterial population from which it then infers information about the entire population through various statistical methods. Even though our isolate panel originates from all African disease foci that have ever yielded positive M. ulcerans cultures, the spatial coverage of disease isolates is moderately restricted to specific geographical areas, which might have confounded our interpretation of the historic spread of African M. ulcerans.
Conclusion
Here we reconstructed the population structure and evolutionary history of African M. ulcerans using the molecular and bioinformatics tools of modern population genomics. The genetic diversity of M. ulcerans proved restricted because of its slow substitution rate coupled with its recent origin. Sequence types appear to be maintained in geographically separated subpopulations that are associated with hydrological drainage areas. Our data suggest that the spread of M. ulcerans across Africa is a relatively modern phenomenon and one that has escalated since the late 19th and the early 20th centuries. Using temporal associations, this work implicates human-induced changes and activities behind the expansion of BU in Africa. We hypothesize that humans with actively infected, openly discharging BU lesions inadvertently contaminated aquatic environments during water contact activities and thus played a role in the spread of the mycobacterium. Our observations on the possible role of humans as potential maintenance reservoir to sustain new BU infections suggests that interventions in a region aimed at reducing the human BU burden will at the same time break the transmission chains within that region. Active case-finding programs, improved disease surveillance, and the early treatment of pre-ulcerative infections with specific antibiotics will decrease the amounts of mycobacteria shed into the environment and may as a result reduce disease transmission. Our findings are supported by the observed decline of BU incidence recorded in some areas which profited from both improved BU surveillance and early treatment (WHO 2015).
Supplementary Material
Acknowledgments
K.V. was supported by a PhD-grant of the Flemish Interuniversity Council—University Development Cooperation (Belgium). B.d.J. and C.M. were supported by the European Research Council-INTERRUPTB starting grant (no. 311725). T.P.S. was supported by a fellowship from the National Health and Medical Research Council of Australia (1105525).
Funding for this work was provided by the Department of Economy, Science and Innovation of the Flemish Government, the Stop Buruli Consortium supported by the UBS Optimus Foundation, and the Fund for Scientific Research Flanders (Belgium) (FWO grant no. G.0321.07N). The computational resources used in this work were provided by the HPC core facility CalcUA and VSC (Flemish Supercomputer Center), funded by the University of Antwerp, the Hercules Foundation and the Flemish Government—department EWI. Aspects of the research in Cameroon and Benin were funded by the Raoul Follereau Fondation France. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank Tanja Stadler for helpful discussions. We thank Pim de Rijk, Wim Mulders, Krista Fissette, Elie Nduwamahoro, and Cécile Uwizeye for their excellent technical assistance.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Authors’ Contributions
Designed research: K.V., M.E., F.P., H.L., T.P.S, and B.J.
Performed research: K.V., K.M., and T.P.S
Contributed new reagents or analytic tools: K.V., C.M., D.A., D.P., S.E., T.S., L.M., E.M., A.C., J.L., A.F., and T.P.S.
Analyzed data: K.V., C.M., and T.P.S.
Wrote the paper: K.V., C.M., T.P.S., K.J., and B.J.
Literature Cited
- Ablordey AS, et al. 2015. Whole genome comparisons suggest random distribution of Mycobacterium ulcerans genotypes in a Buruli ulcer endemic region of Ghana. PLoS Negl Trop Dis. 9:e0003681. doi: 10.1371/journal.pntd.0003681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguiar J. 1997. L'ulcère de Buruli, une maladie mycobactérienne importante et en recrudescence au Bénin. Bull Seances Acad R Sci Outre Mer. 43:325–356. [Google Scholar]
- Andersen FO. 1965. Mycobacterial skin ulcers. Clinical experience. Cent Afr J Med. 11:131–135. [PubMed] [Google Scholar]
- Andrews S. 2015. FastQC: A quality control tool for high throughput sequence data, Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Baele G, et al. 2012. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol Biol Evol. 29:2157–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, et al. 2014. Identifying structural variation in haploid microbial genomes from short-read resequencing data using breseq. BMC Genomics 15:1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolz M, et al. 2015. Locally confined clonal complexes of Mycobacterium ulcerans in two Buruli ulcer endemic regions of Cameroon. PLoS Negl Trop Dis. 9:e0003802.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, et al. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 10:e1003537.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch R, et al. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci U S A. 99:3684–3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruen TC, Philippe H, Bryant D. 2006. A simple and robust statistical test for detecting the presence of recombination. Genetics 172:2665–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas I, et al. 2013. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet. 45:1176–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook A. 1970. Mengo hospital notes, 1897, Makerere Medical School Library. BMJ.. 2:378–379. [Google Scholar]
- Corander J, Marttinen P, Siren J, Tang J. 2008. Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinformatics 9:539.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corander J, Tang J. 2007. Bayesian analysis of population structure based on linked molecular information. Math Biosci. 205:19–31. [DOI] [PubMed] [Google Scholar]
- Croucher NJ, Harris SR, Grad YH, Hanage WP. 2013. Bacterial genomes in epidemiology–present and future. Philos Trans R Soc Lond Ser: B Biol Sci. 368:20120202.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debacker M, et al. 2004. Mycobacterium ulcerans disease: role of age and gender in incidence and morbidity. Trop Med Int Health: TM & IH. 9:1297–1304. [DOI] [PubMed] [Google Scholar]
- Debacker M, Zinsou C, Aguiar J, Meyers WM, Portaels F. 2003. First case of Mycobacterium ulcerans disease (Buruli ulcer) following a human bite. Clin Infect Dis: Off Publ Infect Dis Soc Am. 36:e67–e68. [DOI] [PubMed] [Google Scholar]
- Demangel C, Stinear TP, Cole ST. 2009. Buruli ulcer: reductive evolution enhances pathogenicity of Mycobacterium ulcerans. Nat Rev Microbiol. 7:50–60. [DOI] [PubMed] [Google Scholar]
- Doig KD, et al. 2012. On the origin of Mycobacterium ulcerans, the causative agent of Buruli ulcer. BMC Genomics 13:258.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ho SY, Phillips MJ, Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4:e88.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Nicholls GK, Rodrigo AG, Solomon W. 2002. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics 161:1307–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durnez L, et al. 2009. A comparison of DNA extraction procedures for the detection of Mycobacterium ulcerans, the causative agent of Buruli ulcer, in clinical and environmental specimens. J Microbiol Methods. 76:152–158. [DOI] [PubMed] [Google Scholar]
- Durnez L, et al. 2010. Terrestrial small mammals as reservoirs of Mycobacterium ulcerans in Benin. Appl Environ Microbiol. 76:4574–4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddyani M, et al. 2004. Potential role for fish in transmission of Mycobacterium ulcerans disease (Buruli ulcer): an environmental study. Appl Environ Microbiol. 70:5679–5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddyani M, et al. 2015. A genomic approach to resolving relapse versus reinfection among four cases of Buruli ulcer. PLoS Negl Trop Dis. 9:e0004158.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E, Marth G. 2012. Haplotype-based variant detection from short-read sequencing. arXiv preprint arXiv:1207.3907.
- George KM, et al. 1999. Mycolactone: a polyketide toxin from Mycobacterium ulcerans required for virulence. Science 283:854–857. [DOI] [PubMed] [Google Scholar]
- Giles-Vernick T, Owona-Ntsama J, Landier J, Eyangoh S. 2015. The puzzle of Buruli ulcer transmission, ethno-ecological history and the end of "love" in the Akonolinga district, Cameroon. Soc Sci Med. 129:20–27. [DOI] [PubMed] [Google Scholar]
- Gryseels S, et al. 2012. Amoebae as potential environmental hosts for Mycobacterium ulcerans and other mycobacteria, but doubtful actors in Buruli ulcer epidemiology. PLoS Negl Trop Dis. 6:e1764.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison P, Seeman T. 2014. Nesoni, Available from: https://github.com/Victorian-Bioinformatics-Consortium/nesoni: Victorian Bioinformatics Consortium.
- He M, et al. 2013. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet. 45:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillis DM, Bull JJ. 1993. An empirical-test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol. 42:182–192. [Google Scholar]
- Hodcroft E. 2013. TreeCollapserCL 4. Available from: http://emmahodcroft.com/TreeCollapseCL.html: University of Edinburgh.
- Holt KE, et al. 2012. Shigella sonnei genome sequencing and phylogenetic analysis indicate recent global dissemination from Europe. Nat Genet. 44:1056–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 23:254–267. [DOI] [PubMed] [Google Scholar]
- Jacobsen KH, Padgett JJ. 2010. Risk factors for Mycobacterium ulcerans infection. Int J Infect Dis: IJID: Off Publ Int Soc Infect Dis. 14:e677–e681. [DOI] [PubMed] [Google Scholar]
- Janssens P, Pattyn S, Meyers W, Portaels F. 2005. Buruli ulcer: an historical overview with updating. Bull Seances Acad R Sci Outre Mer. 51:265–299. [Google Scholar]
- Kaser M, et al. 2007. Evolution of two distinct phylogenetic lineages of the emerging human pathogen Mycobacterium ulcerans. BMC Evol Biol. 7:177.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. 2006. Computing Bayes factors using thermodynamic integration. Syst Biol. 55:195–207. [DOI] [PubMed] [Google Scholar]
- Lemey P, Rambaut A, Drummond AJ, Suchard MA. 2009. Bayesian phylogeography finds its roots. PLoS Comput Biol. 5:e1000520.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linz B, et al. 2007. An African origin for the intimate association between humans and Helicobacter pylori. Nature 445:915–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahillon J, Chandler M. 1998. Insertion sequences. Microbiol Mol Biol Rev. 62:725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsollier L, et al. 2007. Impact of Mycobacterium ulcerans biofilm on transmissibility to ecological niches and Buruli ulcer pathogenesis. PLoS Pathog. 3:e62.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marttinen P, et al. 2012. Detection of recombination events in bacterial genomes from large population samples. Nucleic Acids Res. 40:e6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt RW, Benbow ME, Small PLC. 2005. Unraveling an emerging disease associated with disturbed aquatic environments: the case of Buruli ulcer. Front Ecol Environ. 3:323–331. [Google Scholar]
- Meyers WM, et al. 1974a. Distribution of Mycobacterium ulcerans infections in Zaire, including the report of new foci. Ann Soc Belg Med Trop. 54:147–157. [PubMed] [Google Scholar]
- Meyers WM, Shelly WM, Connor DH, Meyers EK. 1974b. Human Mycobacterium ulcerans infections developing at sites of trauma to skin. Am J Trop Med Hygiene. 23:919–923. [DOI] [PubMed] [Google Scholar]
- Monot M, et al. 2009. Comparative genomic and phylogeographic analysis of Mycobacterium leprae. Nat Genet. 41:1282–1289. [DOI] [PubMed] [Google Scholar]
- Moran NA, Plague GR. 2004. Genomic changes following host restriction in bacteria. Curr Opin Genet Dev. 14:627–633. [DOI] [PubMed] [Google Scholar]
- Mortimer TD, Pepperell CS. 2014. Genomic signatures of distributive conjugal transfer among mycobacteria. Genome Biol Evol. 6:2489–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muelder K, Nourou A. 1990. Buruli ulcer in Benin. Lancet 336:1109–1111. [DOI] [PubMed] [Google Scholar]
- Narh CA, et al. 2015. Source tracking Mycobacterium ulcerans infections in the Ashanti region, Ghana. PLoS Negl Trop Dis. 9:e0003437.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakenham T. 1993. The Scramble for Africa: White Man's Conquest of the Dark Continent from 1876 to 1912. New York: Avon Books. [Google Scholar]
- Pearson T, et al. 2004. Phylogenetic discovery bias in Bacillus anthracis using single-nucleotide polymorphisms from whole-genome sequencing. Proc Natl Acad Sci U S A. 101:13536–13541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portaels F, Elsen P, Guimaraes-Peres A, Fonteyne PA, Meyers WM. 1999. Insects in the transmission of Mycobacterium ulcerans infection. Lancet 353:986.. [DOI] [PubMed] [Google Scholar]
- Portaels F, et al. 2008. First cultivation and characterization of Mycobacterium ulcerans from the environment. PLoS Negl Trop Dis. 2:e178.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portaels F, Silva MT, Meyers WM. 2009. Buruli ulcer. Clin Dermatol. 27:291–305. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. 2001. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc Natl Acad Sci U S A. 98:13757–13762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quantum_GIS. 2012. Quantum GIS Geographic Information System Open Source Geospatial Foundation Project.
- R Core Team 2015. R v3.2.0: A language and environment for statistical computing. Available from: http://www.R-project.org/.
- Rambaut A. 2015. FigTree v.1.4.2. Available from: http://tree.bio.ed.ac.uk/software/figtree/.
- Rambaut A. 2008. TreeStat v1.2. Available from: http://tree.bio.ed.ac.uk/software/treestat/.
- Rambaut A, Suchard MA, Xie D, Drummond AJ. 2014. Tracer v1.6, Available from: http://beast.bio.ed.ac.uk/Tracer.
- Ravisse P. 1977. L'ulcère cutané à Mycobacterium ulcerans au Cameroun. Bull Soc Pathol Exotique. 70:109–124. [PubMed] [Google Scholar]
- Ravisse P, Rocques M, Le Bourthe F, Tchuembou C, Menard J. 1975. Une affection méconnue au Cameroun, l'ulcère à Mycobactérie. Med Trop. 471–474. [Google Scholar]
- Rieux A, Balloux F. 2016. Inferences from tip-calibrated phylogenies: a review and a practical guide. Mol Ecol. 25:1911–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röltgen K, Pluschke G. 2015. Mycobacterium ulcerans disease (Buruli ulcer): potential reservoirs and vectors. Curr Clin Microbiol Rep. 2:35–43. [Google Scholar]
- Roltgen K, et al. 2010. Single nucleotide polymorphism typing of Mycobacterium ulcerans reveals focal transmission of Buruli ulcer in a highly endemic region of Ghana. PLoS Negl Trop Dis. 4:e751.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roltgen K, Stinear TP, Pluschke G. 2012. The genome, evolution and diversity of Mycobacterium ulcerans. Infect Genet Evol: J Mol Epidemiol Evol Genet Infect Dis. 12:522–529. [DOI] [PubMed] [Google Scholar]
- Seemann T. 2015. Snippy. Available from: https://github.com/tseemann/snippy.
- Stamatakis A. 2015. The RAxML v8.1.X Manual. Available from: http://sco.h-its.org/exelixis/web/software/raxml/index.html.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinear TP, et al. 2004. Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc Natl Acad Sci U S A. 101:1345–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinear TP, et al. 2007. Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res. 17:192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandelannoote K, et al. 2010. Application of real-time PCR in Ghana, a Buruli ulcer-endemic country, confirms the presence of Mycobacterium ulcerans in the environment. FEMS Microbiol Lett. 304:191–194.20146745 [Google Scholar]
- Vandelannoote K, et al. 2014. Insertion sequence element single nucleotide polymorphism typing provides insights into the population structure and evolution of Mycobacterium ulcerans across Africa. Appl Environ Microbiol. 80:1197–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T, et al. 2008. A landscape-based model for predicting Mycobacterium ulcerans infection (Buruli ulcer disease) presence in Benin, West Africa. EcoHealth 5:69–79. doi: 10.1007/s10393-007-0148-7 [DOI] [PubMed] [Google Scholar]
- Walsh DS, Portaels F, Meyers WM. 2011. Buruli ulcer: advances in understanding Mycobacterium ulcerans infection. Dermatol Clin. 29:1–8. [DOI] [PubMed] [Google Scholar]
- WHO. 2014. Laboratory diagnosis of Buruli ulcer – A manual for health care providers. Geneva: WHO.
- WHO. 2015. WHO meeting on Buruli ulcer Control and Research: Summary report of the control group. In.
- Williamson HR, et al. 2008. Distribution of Mycobacterium ulcerans in Buruli ulcer endemic and non-endemic aquatic sites in Ghana. PLoS Negl Trop Dis. 2:e205.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson HR, et al. 2014. Mycobacterium ulcerans fails to infect through skin abrasions in a Guinea pig infection model: implications for transmission. PLoS Neglected Tropical Diseases 8: e2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip MJ, et al. 2007. Evolution of Mycobacterium ulcerans and other mycolactone-producing mycobacteria from a common Mycobacterium marinum progenitor. J Bacteriol. 189:2021–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.