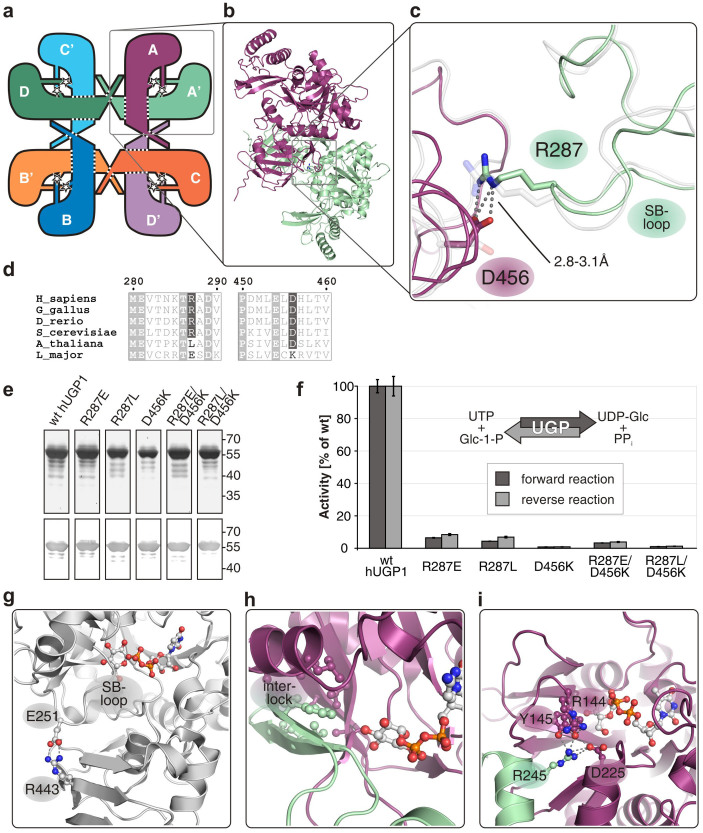
Figure 4. Functional interactions in hUGP and other glucose-activating nucleotidyltransferases.
(a) Schematic representation of the hUGP1 octamer, illustrating oligomer-forming contacts (dotted lines) and functional interactions identified in this study (stars). (b) Side-by-side interactions between two hUGP1 subunits. (c) Functional interaction between R287 (green chain) and D456 (purple chain) of hUGP1 by H-bonds (dashed lines), termed interlock. The corresponding region of superimposed apo-hUGP2 is shown in semi-transparent grey. (d) Sections of the hUGP1 sequence containing residues R287 and D456 (boxed in dark grey) in a multiple sequence alignment of animal, fungal, plant and protozoan UGPs, created with MultAlin and ESPript57,58. Numbering refers to hUGP1. (e) Purified recombinant StrepII-tagged wt hUGP1 and single or double mutants of R287 and D456, analyzed by silver stained SDS-PAGE (top) and Western Blot detecting the StrepII-tag (bottom). (f) In vitro activities of wt and mutant hUGP1 in the forward and reverse reaction. Activitities represent means of quadruplicates ± s.d. and are displayed relative to wt hUGP1 activity. (g) Intramolecular lock mechanism stabilizing the SB-loop in monomeric Leishmania major UGP (PDB ID: 2OEG). Interacting residues and the bound product are highlighted in ball-and-stick representation (see also (h) and (i)). (h) Interlock stabilizing the sugar-binding region in hexameric Salmonella typhi Glc-1-P cytidylyltransferase (PDB ID: 1TZF) by an extended hydrophobic core. (i) Interlock stabilizing the sugar-binding region in tetrameric Pseudomonas aeruginosa Glc-1-P thymidylyltransferase (PDB ID: 1G1L).